Key Points
The propensity of genes to mutate influences the probability of spontaneous reversion of genetic defects in PID.
Abstract
Mutations in T-cell antigen receptor (TCR) subunit genes cause rare immunodeficiency diseases characterized by impaired expression of the TCR at the cell surface and selective T lymphopenia. Here, detailed analyses of spontaneously arising somatic mutations that recover CD247, and thus TCR expression, in a newly identified CD247-deficient patient are described. The recovery of CD247 expression in some patient T cells was associated with both reversion of the inactivating mutation and a variant with a compensating mutation that could reconstitute TCR expression, but not as efficiently as wild-type CD247. Multiple mutations were found in CD247 complementary DNAs (cDNAs) cloned from the patient as well as in cDNA and genomic DNA from other individuals, suggesting that genetic variation in this gene is frequent. Analyses of other genes mutated in primary immunodeficiency diseases (PIDs) where reversions have been described also revealed a higher rate of mutation than that observed for genes mutated in PIDs where revertants have not been identified or control genes. These data support the hypothesis that the occurrence of somatic mutations that may reconstitute genetic defects in PID is related to an increased propensity of those genes to mutate.
Introduction
Spontaneous somatic mutations that repair genetic defects have been documented in a range of primary immunodeficiency diseases (PIDs), including CD247 deficiency,1-4 Wiskott-Aldrich syndrome,5 X-linked IL2RG,6,7 and leukocyte adhesion deficiency,8 among others.9 Although genetic reversion of a deleterious mutation is unlikely, if the mutation confers a selective advantage, these rare clones may expand.9 However, the mechanisms underlying these somatic changes and why they occur in only some PIDs are unclear. Here, a detailed analysis of spontaneously arising somatic mutations that recover CD247, and thus T-cell antigen receptor (TCR) expression, in a CD247-deficient patient is presented. Genetic variation in CD247 is frequent, and this characteristic is shared with other PID genes in which reversion occurs. We suggest that the intrinsic mutability of a gene determines the likelihood of the emergence of somatic revertants on which selection subsequently acts.
Study design
In vitro T-cell culture
Purified peripheral blood mononuclear cells (PBMCs) were stimulated either with irradiated, autologous PBMCs and phytohemagglutinin or with immobilized CD3- and CD28-specific monoclonal antibodies (mAbs). In both cases, interleukin-2 (Peprotech) was added to 50 U/mL.
The study was conducted following the Declaration of Helsinki principles and approved by the institutional research ethics committees involved. All participants, or their guardians, consented to the collection of samples and subsequent analyses.
Sequencing of CD247, CD16A, and FcεRIγ cDNAs
Total RNA and genomic DNA were isolated using the RNeasy Mini kit and DNeasy kit, respectively (QIAGEN). Complementary DNA (cDNA) was synthesized using random hexamers and Superscript II (Life Sciences). Specific exons and full-length cDNAs were amplified using the Expand long-template polymerase chain reaction (PCR) system (Roche) and specific primers (supplemental Table 1, available on the Blood Web site), cloned (CloneJET PCR Cloning Kit), and sequenced (GATC Biotech).
Lentivirus transduction of Ma5.8 cells
CD247 immunoblotting
Cells were lysed in buffer containing 1% Triton X-100/DOC and 0.1% sodium dodecyl sulfate. Then, 30 μg lysate was separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride (Millipore) membrane, and probed with CD247-specific12 and β-actin–specific (Sigma) antibodies.
Flow cytometry
PBMCs were stained with labeled antibodies for CD3, CD56, CD4, CD8 (BioLegend), and TCRαβ (BD Pharmingen). Ma5.8 cells were stained with a phycoerythrin-labeled mAb specific for Vβ3 (BD Pharmingen). For intracellular staining, cells were fixed with paraformaldehyde and permeabilized with saponin before incubation with mAb specific for CD247 (eBioscience), CD3-ε, or -γ (Abcam). Data were acquired using a Gallios cytometer and analyzed using the Kaluza program (Beckman Coulter).
Gene variation analysis
Gene variation data were obtained from the 1000 Genomes Project (http://browser.1000genomes.org). For comparison of missense and synonymous variation, the data were relativized to coding sequence length and plotted as variants per 1000 base pairs. Gene region analysis was carried out by normalizing variants per region to the 5′ untranslated region (UTR), coding sequence, or 3′ UTR length.
Results and discussion
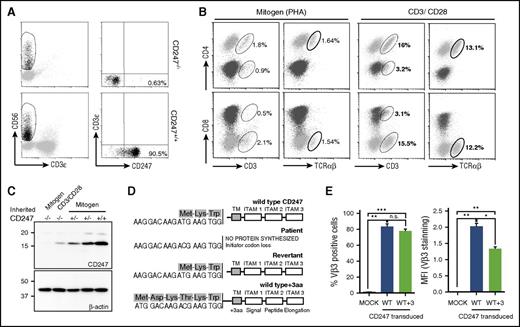
The clinical, genetic, and immunological characteristics of this CD247-deficient patient have been reported previously.3,4 Although most patient T cells were TCR defective, 0.2% of T cells had recovered CD247 expression and normal levels of cell surface CD3ε.4 Rare revertant cells (0.63%) were also present among freshly isolated natural killer (NK) cells (Figure 1A).
Characterization of CD247 revertants. (A) Freshly isolated CD247−/− patient PBMCs were stained, and rare CD247 revertant NK cells were observed. (B-C) Mitogen- and antigen receptor–stimulated PBMCs from a patient were analyzed by flow cytometry (B) and western blot (C) for TCR and CD247 expression. (D-E) Somatic mutants recovering CD247 expression were sequenced (D), and their ability to restore surface TCR expression was analyzed using the CD247-deficient Ma5.8 cell line (E). *P < .05; **P < .01; ***P < .001; n.s. (not significant), P > .05. ITAM, immunoreceptor tyrosine-based activation motif; PHA, phytohemagglutinin; TM, transmembrane; WT, wild-type.
Characterization of CD247 revertants. (A) Freshly isolated CD247−/− patient PBMCs were stained, and rare CD247 revertant NK cells were observed. (B-C) Mitogen- and antigen receptor–stimulated PBMCs from a patient were analyzed by flow cytometry (B) and western blot (C) for TCR and CD247 expression. (D-E) Somatic mutants recovering CD247 expression were sequenced (D), and their ability to restore surface TCR expression was analyzed using the CD247-deficient Ma5.8 cell line (E). *P < .05; **P < .01; ***P < .001; n.s. (not significant), P > .05. ITAM, immunoreceptor tyrosine-based activation motif; PHA, phytohemagglutinin; TM, transmembrane; WT, wild-type.
To study the spontaneous recovery of CD247 expression, patient PBMCs were stimulated with either mitogen (phytohemagglutinin) or immobilized anti-CD3ε/CD28 mAbs. TCR stimulation triggered expansion of these rare CD3high/TCRαβ+ T cells to become ∼18% of the lymphocytes in the culture (Figure 1B), confirming the functionality of these TCR complexes. Western blot analysis demonstrated the recovery of CD247 expression (Figure 1C).
Sequencing of CD247 transcripts cloned from the cultured lymphocytes revealed the presence of the T to C transition eliminating the initiation codon and a revertant, c.2T>C>T, that reconstituted the wild-type initiation codon (Figure 1D). A second mutation, c.-8A>T, producing a new methionine codon permitting synthesis of a mutant CD247 with a 3-amino-acid extension of the signal peptide (WT+3) (Figure 1D), was also identified. Transfection of cell-surface TCR-negative Ma5.8 cells11 with wild-type CD247 or the WT+3 variant restored TCR expression (Figure 1E). However, the MFI for TCR staining was significantly lower for cells expressing the WT+3 variant (Figure 1E).
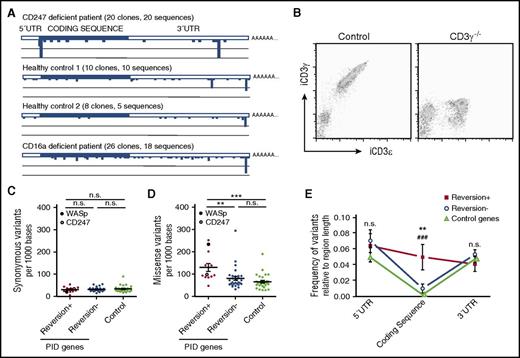
The sequencing analysis also revealed that mutations occurred not only around the initiation codon but also throughout the CD247 transcript so that 15 sequences were found among 20 full-length cDNA clones sequenced (supplemental Table 2). In contrast, no variation was noted in transcripts of the FcγRIIIA and FcεR1γ genes (data not shown) also encoded on chromosome 1. Multiple, distinct CD247 cDNA clones were also isolated from 3 other individuals (Figure 2A; supplemental Table 2), but again, no sequence variation was noted for FcεR1γ. Sequence analysis of CD247 exons amplified from genomic DNA isolated from purified granulocytes confirmed the frequent sequence variation in CD247 (supplemental Table 3). In control experiments, no revertants were noted in freshly isolated T cells of CD3γ-deficient patients13 (Figure 2B). No revertant clones were identified after sequencing CD3γ cDNA clones prepared from in vitro–activated T cells; indeed, the majority of sequences were invariant, with only 3 single-nucleotide variants identified (data not shown).
Gene variation and reversion probability in PIDs. (A) CD247 sequence analysis of samples from the patient and controls revealed high genetic variability. (B) Freshly isolated PBMCs from a CD3γ−/− patient and control were stained and analyzed by flow cytometry. (C-E) Analysis of data from the 1000 Genomes Project revealed no differences in the occurrence of synonymous mutations (C), but an increased frequency of missense mutations (D) in the coding region (E) was observed for PID genes where reversion has been described compared with PID genes with no reversion described or control genes. CD247 and WASp are specifically indicated in panels C and D. Statistical significance was calculated using a 1-way analysis of variance for synonymous and missense variation rates and a 2-way analysis of variance for variation vs gene region analysis comparing the 3 different groups of genes (**P < .01; ***P < .001; n.s., P > .05).
Gene variation and reversion probability in PIDs. (A) CD247 sequence analysis of samples from the patient and controls revealed high genetic variability. (B) Freshly isolated PBMCs from a CD3γ−/− patient and control were stained and analyzed by flow cytometry. (C-E) Analysis of data from the 1000 Genomes Project revealed no differences in the occurrence of synonymous mutations (C), but an increased frequency of missense mutations (D) in the coding region (E) was observed for PID genes where reversion has been described compared with PID genes with no reversion described or control genes. CD247 and WASp are specifically indicated in panels C and D. Statistical significance was calculated using a 1-way analysis of variance for synonymous and missense variation rates and a 2-way analysis of variance for variation vs gene region analysis comparing the 3 different groups of genes (**P < .01; ***P < .001; n.s., P > .05).
Previous molecular characterization of revertant cells has generally revealed one or, rarely, a few revertant genotypes.9 However, these observations likely underestimate genotypic diversity, since direct sequence analysis of PCR-amplified DNA will only identify genotypes that have undergone sufficient clonal expansion to reach a detectable frequency. Similar considerations undermine the suggestion that CD247 was not a mutational hot spot, since somatic mutants were only found in patient T cells.1 CD247 is only expressed in T and NK cells that can undergo sufficient clonal expansion for the mutant genotype to be detectable. Our data show that CD247 mutants are frequent, although functional consequences are hard to quantify, since these mutations likely occur in single cells in heterozygosis.
Consistent with the idea that genetic diversity in PID might be greater than often appreciated, direct PCR analysis of purified WASp-expressing T cells from a WASp-deficient patient identified 1 mutation, whereas 25 putative revertant genotypes were identified when 45 allospecific T-cell clones were analyzed.5 This diversity of WASp sequences in revertant T cells was confirmed by high-resolution electrophoretic analysis of WASp RNA from another WASp-deficient patient.14
Thus, the occurrence of revertant or compensating mutations in certain PIDs might reflect a more general tendency of those genes to mutate (ie, more mutation means more chance of a reversion/compensation event that might confer a selective advantage). To test this hypothesis, we compared the genetic variability of a set of control genes, PID genes where reversion has been described, and PID genes without revertants. Here, it should be remembered that these are rare diseases, so it is possible that as more patients are described in detail, the definition of some PIDs as “nonrevertant” might change. This analysis is fundamentally different from those above, since genetic variability within a population is being studied, whereas Figure 2A reports CD247 variation within individuals. The rate of missense, but not synonymous, mutations observed in PID genes where reversion has been described was significantly higher than that found for nonrevertants or control genes (Figure 2C-D). These observations support the hypothesis that certain PID genes are more mutable than others and that an increased mutation rate makes it more likely that reversion or compensating mutations may occur. The nature of the gene mutated and the degree of recovery of function will affect these data, since they will influence the possibilities of clonal expansion and thus detection. However, the possible impact of these mutations on gene function, as assessed by the Gene Damage Index algorithm,15 was not significantly different (supplemental Table 4). Since genes important for lymphocyte growth are found among both PID genes with revertant and nonrevertant PID genes (supplemental Table 4), we suggest that the frequency of mutation is a key first influence on the emergence of candidate revertants that subsequently are the objects of selection that may lead to clonal expansion. Interestingly, variation in the coding sequence was significantly greater for revertant genes than for nonrevertant or control genes but similar at UTR flanking regions (Figure 2E). The genetics underlying the intrinsic mutability of PID genes where reversion occurs is unknown. The presence of CpG islands was somewhat more frequent in revertant than nonrevertant PID genes (supplemental Table 4), but it is also possible that local chromatin structure and accessibility for DNA repair could influence mutation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Hisse van Santen and Balbino Alarcón for gifts of CD247-specific antibody and Ma5.8 cells and Almudena R. Ramiro for critical reading of the manuscript.
This work was supported in part by grants SAF2014-58752-R (H.T.R.), SAF2014-54708-R (J.R.R.), and SAF2015-69169-R (M.V.-G.) from the Ministerio de Economia y Competitividad and by PhD studentships to A.B.-M. (MINECO SVP-2014-068263) and A.P.-P. (National Secretary of Higher Education, Science, Technology and Innovation, SENESCYT, Ecuador).
Authorship
Contribution: A.B.-M. performed research, analyzed data, and wrote the paper; A.P.-P., M.A.-L., and D.D. performed research and analyzed data; M.V.-G. and J.R.R. analyzed data and wrote the paper; C.A. contributed vital new reagents or analytical tools; A.I. contributed vital new reagents or analytical tools and wrote the paper; and H.T.R. designed research, performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hugh T. Reyburn, CNB-CSIC, Darwin 3, Campus de CantoBlanco, 28049 Madrid, Spain; e-mail: htreyburn@cnb.csic.es.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal