Key Points
Plasma-derived exosomes from patients with CLL exhibit different protein cargo compositions depending on disease status and progression.
S100-A9 protein is overexpressed and S100-A9 cargo in exosomes activates NF-κB pathway in patients with CLL during disease progression.
Abstract
Chronic lymphocytic leukemia (CLL) is an incurable disease characterized by accumulation of clonal B lymphocytes, resulting from a complex balance between cell proliferation and apoptotic death. Continuous crosstalk between cancer cells and local/distant host environment is required for effective tumor growth. Among the main actors of this dynamic interplay between tumoral cells and their microenvironment are the nano-sized vesicles called exosomes. Emerging evidence indicates that secretion, composition, and functional capacity of exosomes are altered as tumors progress to an aggressive phenotype. In CLL, no data exist exploring the specific changes in the proteomic profile of plasma-derived exosomes from patients during disease evolution. We hereby report for the first time different proteomic profiles of plasma exosomes, both between indolent and progressive CLLs as well as within the individual patients at the onset of disease and during its progression. Next, we focus on the changes of the exosome protein cargoes, which are found exclusively in patients with progressive CLL after disease progression. The alterations in the proteomic cargoes underline different networks specific for leukemia progression related to inflammation, oxidative stress, and NF-κB and phosphatidylinositol 3-kinase/AKT pathway activation. Finally, our results suggest a preponderant role for the protein S100-A9 as an activator of the NFκB pathway during CLL progression and suggest that the leukemic clone can generate an autoactivation loop through S100-A9 expression, NF-κB activation, and exosome secretion. Collectively, our data propose a new pathway for NF-κB activation in CLL and highlight the importance of exosomes as extracellular mediators promoting tumor progression in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) develops through accumulation of malignant B cells that circulate in the blood and are continuously supported by microenvironment signals within the bone marrow and secondary lymphoid organs.1 Exchange between primary tumor cells and their microenvironment represents bidirectional crosstalk, in which the tumor clone is not only being supported by the microenvironment but also modulating it to maintain and promote its own survival.2,3
Among the main actors in the dynamic and reciprocal interplay between tumor cells and their microenvironment are extracellular vesicles (EVs).4 They represent an important mode of intercellular communication, serving as vehicles for transfer between cellular membranes, proteins, lipids, and RNA. EVs include microvesicles (MVs), which shed directly from the plasma membrane with a diameter of 100 to 1000 nm,5 and exosomes, which are conformed by nano-sized vesicles of endosomal origin of 40 to 100 nm in diameter.6 Similar to MVs, exosomes are directly associated with oncogenic reprogramming,7 tumor progression,8 metastasis,9 and therapy refractoriness.10
In CLL, the total number of MVs in plasma is greater than in healthy donors, and in vitro studies have demonstrated that MVs could mediate AKT activation in bone marrow stromal cells.11 Regarding CLL exosomes, recent work has demonstrated that they could induce an inflammatory phenotype through a transition of stromal cells into cancer-associated fibroblasts and activation of key signaling pathways such as AKT and/or NF-κB.12 Although microRNAs and proteomic profiles of exosomes have been described,12,13 proteomic analysis of primary exosomes throughout CLL evolution has not been fully performed. Furthermore, the direct implications of changes occurring in exosome protein cargoes throughout CLL evolution and their role during disease progression remain unknown.
We hereby report a comprehensive analysis involving the proteomic characterization of exosomes from patients with indolent or progressive CLL at the onset of disease and during disease evolution. We examined whether there is a differential proteomic profile from exosomes throughout CLL evolution and focused on the different pathways and proteins that could be implicated in disease progression. We found that S100-A9 protein is only present in exosomes of patients with progressive disease at the time of progression and that this protein is specifically overexpressed by leukemic cells at this stage. Finally, we demonstrate that exosomes carrying S100-A9 are able to activate the NF-κB pathway in primary CLL cells, suggesting that the leukemic clone could generate an autoactivation loop through S100-A9 expression, exosome secretion, and NF-κB activation.
Together, these results show the importance of exosomes in cell-to-cell communication during CLL evolution and suggest that exosomes with S100-A9 protein cargo could be one of the mechanisms responsible for the activation of the NF-κB pathway during disease progression.
Materials and methods
Clinical samples
Plasma and peripheral blood mononuclear cells (PBMCs) were collected in a prospective study from 34 patients with typical CLL diagnoses. Five age-matched healthy donors (HDs) were included as control samples. Patients were segregated into 2 groups: (1) progressive disease, defined by lymphocyte doubling time <6 months, treatment required within 3 years, activation-induced cytidine deaminase/lipoprotein lipase expression, and >2% of CLL B cells with ongoing class-switch recombination process in peripheral blood (PB), as described by Palacios et al,14 and (2) indolent disease, defined by lymphocyte doubling time >4 years, no treatment required after 4 years, absence in PB (<1%) of CLL B cells with ongoing class-switch recombination process to immunoglobulin G (IgG), and absence of activation-induced cytidine deaminase expression. All samples were obtained at disease onset and after 4 years during disease evolution or after treatment in those with progressive disease. All patients were observed at Hospital Maciel and at the Haematology Department of Clinical Hospital in Montevideo. They provided informed consent according to the ethical regulations of Uruguay and the Helsinki Declaration.
Exosome isolation
Patients’ plasmas were diluted 1:2 in phosphate-buffered saline and centrifuged (10 minutes at 400 g, then 20 minutes at 2000 g) to remove cell debris and larger vesicles. Next, centrifugation at 10 000 g for 60 minutes at 4C° was performed to separate MVs. Exosomes were isolated by ultracentrifugation (120 minutes at 100 000 g at 4°C) followed by floatation on Optiprep cushion (Axis-Shield; 17%) for 120 minutes at 100 000 g at 4°C to remove nonexosomal protein complexes. After washing, exosomes were suspended in phosphate-buffered saline and subjected to NanoSight analysis (Malvern Instruments, Inc., Westborough, MA).
Proteomic analysis
Peptides were separated by nano–high-performance liquid chromatography (EASY 1000; Thermo Fisher) fitted with a reverse-phase column (50 cm × 75 µm ID, PepMap RSLC C18; Thermo Fisher) using a linear gradient (acetonitrile, 0.1%; formic acid, 5%-55% in 75 minutes). Peptide analysis was carried out via an LTQ Velos (Thermo Fisher) in a data-dependent acquisition mode (top 10) with a dynamic exclusion list of 45 seconds. Data analysis was performed using PatternLab for proteomics software (version 3.2.0.3).15,16 Raw files were searched against a Homo sapiens target-decoy database using the Comet search engine (PatternLab). The approximate area proportional Venn diagram module was used to perform comparisons between different progression times and determine proteins uniquely identified in each situation.16 Exosomal protein data sets were subjected to functional enrichment analyses using FunRich software.17 The mass spectrometry proteomic data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD005941.
Flow cytometry, western blot, confocal microscopy, and reverse-transcription polymerase chain reaction
Isolation of PBMCs from patients with CLL and HDs, as well as cytometry analysis (flow cytometry) and immunoblot, was performed as described.18,19 Antibodies used in this work are described in supplemental Table 1, available on the Blood Web site. Flow cytometry analysis was performed with R/Bioconductor20 packages or FlowJo (Treestar, Ashland, OR) or Summit software (Beckman Coulter, CA). Microscopy analysis was performed as previously described,21 with antibodies listed in supplemental Table 1. RNA isolation and reverse-transcription polymerase chain reaction were carried out as described by Oppezzo et al.22 Primers used for the amplification of extracellular matrix metalloprotease inducer (EMMPRIN) isoforms and the full-length S100-A9 are provided in supplemental Table 2.
In vitro analysis
Purification of CD19+ cells from patients with CLL and activation with anti-IgM (Jackson ImmunoResearch) were performed as described by Moreno et al.23 Exosome activation on CLL cells was performed with PBMCs (5 × 106) cultured in RPMI 1640, 10% fetal bovine serum and incubated for 90 minutes with exosomes (30 µg/mL total protein). For blocking experiments, exosomes were preincubated with 1 μg/mL of anti-human S100-A9 (R&D Systems, Minneapolis, MN) or Human IgG Isotype Control (Thermo Fisher) for 1 hour at 4°C. Recombinant human S100-A9 protein (rhS100-A9; MyBiosource, San Diego, CA) was incubated with CLL PBMCs as described in supplemental Table 1.
Statistical analysis
Results are expressed as the mean ± standard error of the mean of at least 3 independent experiments. Differences between groups were analyzed using the Mann-Whitney test or 1-way analysis of variance (Student t test); P < .05 was considered statistically significant. Analyses were performed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA). For proteomic assays, false-discovery rate correction was performed using GeneSpring GX11.0.2 software (Agilent Technologies, Santa Clara, CA); cutoff of enrichment analyses in FunRich software was kept as default (P < .05) after Bonferroni correction.
Results
Characterization of patients with CLL and plasma-derived exosomes
To determine the clinical relevance of circulating exosomes in patients with CLL, we isolated and characterized exosomes from the plasma of a working cohort composed of: (1) group A, comprising 5 patients with progressive disease, with samples obtained at the time of diagnosis during stable disease (Prog-dt) and later during disease progression (Prog-ddp) but before treatment, and (2) group B, consisting of 5 patients with indolent disease, with samples obtained at diagnosis (Ind-dt) and after 4 years of follow-up without disease evolution (Ind-4years). Rationale of the study, clinical statement, and molecular characterization of patients with CLL are provided in Table 1.
Clinical and molecular characterization of patients with CLL
| Study rationale . | Patient No. . | CLL samples at time of disease onset (subgroups Prog-dt and Ind-dt) . | CLL samples during disease progression or after 4 y of follow-up (subgroups Prog-ddp and Ind-4years) . | TFT, months . | IgVH status‡ . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Binet stage . | LC, × 103/μl . | Clinical disease status . | FISH . | AID/LPL* . | CSR, %† . | Binet stage . | LC, × 103/μl . | Clinical disease status . | FISH . | AID/LPL* . | CSR, %† . | ||||
| Study cohort (progressive patients) | 1 | A | 19.0 | Ind. | Normal | (+/+) | 2.5 | B | 74.00 | Prog. | del17p | (+/+) | 2.0 | 18 | UM |
| 2 | A | 28.0 | Ind. | del11q | (+/+) | 2.1 | B | 162.0 | Prog. | del11q | (+/+) | 2.9 | 28 | UM | |
| 3 | A | 23.00 | Ind. | del13q/del11q | (+/+) | 2.8 | C | 120.0 | Prog. | del13q/del11q | (+/+) | 3.1 | 32 | UM | |
| 4 | A | 35.00 | Ind. | del11q | (+/+) | 4.1 | C | 164.0 | Prog. | del11q | (+/+) | 5.5 | 14 | UM | |
| 5 | A | 15.90 | Ind. | Normal | (+/+) | 3.8 | C | 220.0 | Prog. | Normal | (+/+) | 4.2 | 9 | UM | |
| Study cohort (indolent patients) | 6 | A | 27.70 | Ind. | Normal | (−/−) | 1.2 | A | 19.64 | Ind. | Normal | (−/−) | 0.9 | N/T | Mut |
| 7 | A | 24.60 | Ind. | Normal | (−/−) | 0.02 | A | 34.55 | Ind. | Normal | (−/−) | 0.5 | N/T | Mut | |
| 8 | A | 14.70 | Ind. | del13q | (−/−) | 1.1 | A | 12.50 | Ind. | del13q | (+/−) | 1.2 | N/T | Mut | |
| 9 | A | 8.25 | Ind. | del13q | (−/−) | 0.8 | A | 14 490 | Ind. | del13q | (−/−) | 0.3 | N/T | Mut | |
| 10 | A | 18.00 | Ind. | Normal | (−/−) | 1.5 | A | 20.00 | Ind. | Normal | (−/−) | 1.8 | N/T | Mut | |
| Validation cohort (progressive patients) | 11 | A | 26.20 | Ind. | del13q | (+/−) | 1.5 | C | 183.0 | Prog. | del13q | (+/−) | 2.2 | 33 | UM |
| 12 | A | 19.45 | Ind. | Normal | (−/+) | 0.5 | B | 112.0 | Prog. | del17p | (+/+) | 2.5 | 35 | UM | |
| 13 | A | 40.20 | Ind. | del13q | (+/+) | 1.2 | B | 98.00 | Prog. | del11q | (+/+) | 2.2 | 31 | UM | |
| 14 | A | 18.30 | Ind. | del13q/del11q | (+/+) | 1.6 | C | 178.00 | Prog. | del13q/del11q | (+/+) | 3.1 | 6 | UM | |
| 15 | A | 20.26 | Ind. | del17p | (−/+) | 1.8 | B | 154.00 | Prog. | del17p | (+/+) | 2.3 | 40 | UM | |
| 16 | A | 10.58 | Ind. | del11q | (+/+) | 0.5 | B | 120.0 | Prog. | del11q | (+/+) | 1.8 | 10 | UM | |
| 17 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 77.00 | Prog. | del11q | (+/+) | 3.6 | 9 | UM | |
| 18 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | C | 161.00 | Prog. | del13q | (+/−) | 2.9 | 11 | UM | |
| 19 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 72.0 | Prog. | del11q | (+/+) | 4.4 | 38 | UM | |
| 20 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | C | 900.0 | Prog. | Normal | (+/+) | 1.2 | 8 | UM | |
| 21 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 110.0 | Prog. | del13q | (−/+) | 4.4 | 12 | UM | |
| 22 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | C | 97.00 | Prog. | Normal | (+/+) | 2.5 | 26 | UM | |
| 23 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 65.00 | Prog. | Normal | (+/+) | 1.5 | 24 | UM | |
| Validation cohort (indolent patients) | 24 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 15.60 | Ind. | Normal | (−/+) | 0.6 | 130 | Mut |
| 25 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 8.90 | Ind. | tris12 | (−/−) | 0.5 | N/T | Mut | |
| 26 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 9.92 | Ind. | Normal | (−/−) | 0.07 | N/T | Mut | |
| 27 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 15.00 | Ind. | Normal | (−/−) | 0.05 | N/T | Mut | |
| 28 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 7.00 | Ind. | del13q | (−/+) | 1.5 | N/T | Mut | |
| 29 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 11.60 | Ind. | del13q | (+/−) | 1.1 | N/T | Mut | |
| 30 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 14.70 | Ind. | tri12 | (−/−) | 2.1 | 126 | Mut | |
| 31 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 22.20 | Ind. | Normal | (−/+) | 1.8 | N/T | Mut | |
| 32 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 11.60 | Ind. | Normal | (−/−) | 0.6 | N/T | Mut | |
| 33 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 17.50 | Ind. | Normal | (−/−) | 0.7 | N/T | Mut | |
| 34 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 19.90 | Ind. | del13q | (−/−) | 1.1 | N/T | Mut | |
| Study rationale . | Patient No. . | CLL samples at time of disease onset (subgroups Prog-dt and Ind-dt) . | CLL samples during disease progression or after 4 y of follow-up (subgroups Prog-ddp and Ind-4years) . | TFT, months . | IgVH status‡ . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Binet stage . | LC, × 103/μl . | Clinical disease status . | FISH . | AID/LPL* . | CSR, %† . | Binet stage . | LC, × 103/μl . | Clinical disease status . | FISH . | AID/LPL* . | CSR, %† . | ||||
| Study cohort (progressive patients) | 1 | A | 19.0 | Ind. | Normal | (+/+) | 2.5 | B | 74.00 | Prog. | del17p | (+/+) | 2.0 | 18 | UM |
| 2 | A | 28.0 | Ind. | del11q | (+/+) | 2.1 | B | 162.0 | Prog. | del11q | (+/+) | 2.9 | 28 | UM | |
| 3 | A | 23.00 | Ind. | del13q/del11q | (+/+) | 2.8 | C | 120.0 | Prog. | del13q/del11q | (+/+) | 3.1 | 32 | UM | |
| 4 | A | 35.00 | Ind. | del11q | (+/+) | 4.1 | C | 164.0 | Prog. | del11q | (+/+) | 5.5 | 14 | UM | |
| 5 | A | 15.90 | Ind. | Normal | (+/+) | 3.8 | C | 220.0 | Prog. | Normal | (+/+) | 4.2 | 9 | UM | |
| Study cohort (indolent patients) | 6 | A | 27.70 | Ind. | Normal | (−/−) | 1.2 | A | 19.64 | Ind. | Normal | (−/−) | 0.9 | N/T | Mut |
| 7 | A | 24.60 | Ind. | Normal | (−/−) | 0.02 | A | 34.55 | Ind. | Normal | (−/−) | 0.5 | N/T | Mut | |
| 8 | A | 14.70 | Ind. | del13q | (−/−) | 1.1 | A | 12.50 | Ind. | del13q | (+/−) | 1.2 | N/T | Mut | |
| 9 | A | 8.25 | Ind. | del13q | (−/−) | 0.8 | A | 14 490 | Ind. | del13q | (−/−) | 0.3 | N/T | Mut | |
| 10 | A | 18.00 | Ind. | Normal | (−/−) | 1.5 | A | 20.00 | Ind. | Normal | (−/−) | 1.8 | N/T | Mut | |
| Validation cohort (progressive patients) | 11 | A | 26.20 | Ind. | del13q | (+/−) | 1.5 | C | 183.0 | Prog. | del13q | (+/−) | 2.2 | 33 | UM |
| 12 | A | 19.45 | Ind. | Normal | (−/+) | 0.5 | B | 112.0 | Prog. | del17p | (+/+) | 2.5 | 35 | UM | |
| 13 | A | 40.20 | Ind. | del13q | (+/+) | 1.2 | B | 98.00 | Prog. | del11q | (+/+) | 2.2 | 31 | UM | |
| 14 | A | 18.30 | Ind. | del13q/del11q | (+/+) | 1.6 | C | 178.00 | Prog. | del13q/del11q | (+/+) | 3.1 | 6 | UM | |
| 15 | A | 20.26 | Ind. | del17p | (−/+) | 1.8 | B | 154.00 | Prog. | del17p | (+/+) | 2.3 | 40 | UM | |
| 16 | A | 10.58 | Ind. | del11q | (+/+) | 0.5 | B | 120.0 | Prog. | del11q | (+/+) | 1.8 | 10 | UM | |
| 17 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 77.00 | Prog. | del11q | (+/+) | 3.6 | 9 | UM | |
| 18 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | C | 161.00 | Prog. | del13q | (+/−) | 2.9 | 11 | UM | |
| 19 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 72.0 | Prog. | del11q | (+/+) | 4.4 | 38 | UM | |
| 20 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | C | 900.0 | Prog. | Normal | (+/+) | 1.2 | 8 | UM | |
| 21 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 110.0 | Prog. | del13q | (−/+) | 4.4 | 12 | UM | |
| 22 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | C | 97.00 | Prog. | Normal | (+/+) | 2.5 | 26 | UM | |
| 23 | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | N/A(1) | B | 65.00 | Prog. | Normal | (+/+) | 1.5 | 24 | UM | |
| Validation cohort (indolent patients) | 24 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 15.60 | Ind. | Normal | (−/+) | 0.6 | 130 | Mut |
| 25 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 8.90 | Ind. | tris12 | (−/−) | 0.5 | N/T | Mut | |
| 26 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 9.92 | Ind. | Normal | (−/−) | 0.07 | N/T | Mut | |
| 27 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 15.00 | Ind. | Normal | (−/−) | 0.05 | N/T | Mut | |
| 28 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 7.00 | Ind. | del13q | (−/+) | 1.5 | N/T | Mut | |
| 29 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 11.60 | Ind. | del13q | (+/−) | 1.1 | N/T | Mut | |
| 30 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 14.70 | Ind. | tri12 | (−/−) | 2.1 | 126 | Mut | |
| 31 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 22.20 | Ind. | Normal | (−/+) | 1.8 | N/T | Mut | |
| 32 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 11.60 | Ind. | Normal | (−/−) | 0.6 | N/T | Mut | |
| 33 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 17.50 | Ind. | Normal | (−/−) | 0.7 | N/T | Mut | |
| 34 | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | N/A(2) | A | 19.90 | Ind. | del13q | (−/−) | 1.1 | N/T | Mut | |
AID, activation-induced cytidine deaminase; CSR, class-switch recombination; FISH, fluorescence in situ hybridization; LC, lymphocyte count; LPL, lipoprotein lipase; Mut, mutated; N/A(1), not applicable (refers to patients with progressive disease whose samples were not collected at disease onset); N/A(2), not applicable (refers to analyzed samples collected from indolent patients after 4 years of follow-up); N/T, not treated (refers to patients who did not receive any treatment at 4-year follow-up); TFT, time from initial diagnosis to first treatment for clinical progression; UM, unmutated.
AID and LPL expression were assessed by quantitative polymerase chain reaction, as described by Prieto et al.21
Percentage of proliferative fraction expressing IgM+/IgG+ evaluated by cytometry studies, as described by Palacios et al.14
≤ 2% difference from germline gene defined UM patients; ≥ 2% difference defined Mut patients.
Isolated particles presented bona fide characteristics of exosomes, with size ranging from 40 to 100 nm and a typical profile confirmed by NanoSight and electron microscopy (Figure 1A). Additionally, different protein migration patterns were visualized between exosomes and whole-cell lysate of the same patients, whereas classical exosome markers such as CD9, HSP-70, and Lamp-124 were enriched in the exosome fraction. In turn, cytochrome C was absent, excluding contamination with apoptotic bodies (Figure 1B). Component enrichment analysis of the different exosome fractions showed mainly exosome proteins, which indicates that the procedures used for isolation and purification are reproducible and reliable (Figure 1C).
Characterization and proteomic analysis of CLL plasma-derived exosomes. (A) Electron microscopy and NanoSight analysis of exosomes (Exo) purified from CLL plasmas. Comparative electron micrographs of MVs and exosomes from the same patient are depicted. Blue numbers indicate size of main peaks. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot characterization of plasma-derived exosomes. After protein quantification, exosomes were lysed in Laemmli buffer to perform SDS-PAGE and immunoblot. Different migration profiles in SDS-PAGE of exosome fractions and immunoblot with specific monoclonal antibodies against typical exosome proteins were used to validate the quality of our isolation technique. The proteins identified were tetraspanin CD9, heat shock protein HSP-70, and lysosomal marker LAMP-1. These were compared with the plasma of the same patient at the same disease time. Cytochrome C was also evaluated comparing whole-cell lysate (WCL) with plasma-derived exosomes of the same patient. (C) Subcellular localization of the proteins present in the plasma-derived exosomes purified of the different subgroups was analyzed by FunRich software. MWM, molecular-weight marker.
Characterization and proteomic analysis of CLL plasma-derived exosomes. (A) Electron microscopy and NanoSight analysis of exosomes (Exo) purified from CLL plasmas. Comparative electron micrographs of MVs and exosomes from the same patient are depicted. Blue numbers indicate size of main peaks. (B) Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot characterization of plasma-derived exosomes. After protein quantification, exosomes were lysed in Laemmli buffer to perform SDS-PAGE and immunoblot. Different migration profiles in SDS-PAGE of exosome fractions and immunoblot with specific monoclonal antibodies against typical exosome proteins were used to validate the quality of our isolation technique. The proteins identified were tetraspanin CD9, heat shock protein HSP-70, and lysosomal marker LAMP-1. These were compared with the plasma of the same patient at the same disease time. Cytochrome C was also evaluated comparing whole-cell lysate (WCL) with plasma-derived exosomes of the same patient. (C) Subcellular localization of the proteins present in the plasma-derived exosomes purified of the different subgroups was analyzed by FunRich software. MWM, molecular-weight marker.
Proteomic profiling of plasma-derived exosomes during CLL evolution
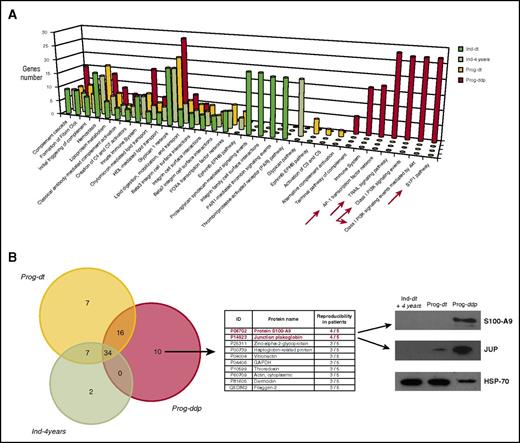
Considering all patients and conditions, liquid chromatography–tandem mass spectrometry analysis of CLL exosomes resulted in the identification of 138 proteins in group A and 136 proteins in group B, with at least 2 mapping peptides per sequence. Among the 138 proteins in the progressive group, 99 (62%) were shared by the 2 subgroups (Prog-dt and Prog-ddp), whereas 39 proteins were differentially expressed: 10 in Prog-dt and 29 in Prog-ddp. Regarding indolent CLLs, among the 136 proteins, 84 (61%) were shared between both subgroups (Ind-dt and Ind-4years), and 52 proteins were differentially identified: 12 in Ind-dt and 40 in Ind-4years (supplemental Data 1, including tables and Venn diagram). The protein-protein interaction (PPI) network offers a conceptual framework to better understand the functional organization of the proteome and, in this case, to deepen the insight into the protein cargoes of plasma exosomes during CLL evolution. Selection of the 20 most significant PPI networks from each subgroup (Ind-dt, Ind-4years, Prog-dt, and Prog-ddp) and further comparison among them allowed us to highlight a total of 32 different biological pathways (Figure 2A; supplemental Data 2). Many of these pathways were shared among progressive and indolent CLLs, but some of them were found exclusively within a determined subgroup (Figure 2A). Focusing on the PPI networks derived from the proteins present in exosomes during disease progression (Prog-ddp subgroup), we found interesting pathways associated with inflammation and cancer progression, such as S1P1 pathway, class I PI3K signaling events, tumor necrosis factor–related apoptosis-inducing ligand signaling pathway, and AP-1 transcription factor network (Figure 2A).
Proteomic analysis of plasma-derived exosomes during CLL evolution. (A) Molecular networks associated with proteins identified in plasma-derived exosomes during CLL evolution. The PPI networks were elucidated using the interaction model of FunRich software with the Vesiclepedia database, which focuses on statistically significant enriched genes and particular biological pathways in each subgroup. Different and specific networks according to the number of genes associated with the functions in the Prog-ddp subgroup are marked by red arrows: (1) Leukemic cell infiltration of secondary lymphoid organs, highlighted by genes related to the receptor sphingosine 1-phosphate (S1P1) pathway (P = 2.24e−4); (2) tumor proliferation network, involving genes of phosphatidylinositol 3-kinase (PI3K)/AKT kinase pathway (P = 2.24e−4); (3) survival and apoptosis network, involving genes implicated in the tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) pathway; and (4) inflammation and oxidative stress networks, with genes involved in the AP-1 transcription factor pathway (P = 1.43e−4). (B) Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis of CLL exosome proteins. LC-MS/MS data analysis was performed in accordance with the recently published PatternLab for proteomics 4.0 software (http://www.patternlabforproteomics.org) protocol for data analysis.16 The PatternLab approximately area-proportional Venn diagram module was used for pinpointing proteins exclusively identified in 1 biological condition. For the exosome protein-enriched sample, the analysis only considered proteins present in at least 3 of 5 patients for each biological condition. Junction plakoglobin (JUP) and S100-A9 (proteins in red font) were confirmed by specific monoclonal antibodies in immunoblot analysis. Plasma-derived exosomes were obtained from 10 patients (20 samples) corresponding to the 4 different subgroups (5 patients each) and subjected to 12% polyacrylamide gel (SDS-PAGE) electrophoresis. Representative immunoblots of both proteins at the different disease times are depicted. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDL, high-density lipoprotein.
Proteomic analysis of plasma-derived exosomes during CLL evolution. (A) Molecular networks associated with proteins identified in plasma-derived exosomes during CLL evolution. The PPI networks were elucidated using the interaction model of FunRich software with the Vesiclepedia database, which focuses on statistically significant enriched genes and particular biological pathways in each subgroup. Different and specific networks according to the number of genes associated with the functions in the Prog-ddp subgroup are marked by red arrows: (1) Leukemic cell infiltration of secondary lymphoid organs, highlighted by genes related to the receptor sphingosine 1-phosphate (S1P1) pathway (P = 2.24e−4); (2) tumor proliferation network, involving genes of phosphatidylinositol 3-kinase (PI3K)/AKT kinase pathway (P = 2.24e−4); (3) survival and apoptosis network, involving genes implicated in the tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) pathway; and (4) inflammation and oxidative stress networks, with genes involved in the AP-1 transcription factor pathway (P = 1.43e−4). (B) Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis of CLL exosome proteins. LC-MS/MS data analysis was performed in accordance with the recently published PatternLab for proteomics 4.0 software (http://www.patternlabforproteomics.org) protocol for data analysis.16 The PatternLab approximately area-proportional Venn diagram module was used for pinpointing proteins exclusively identified in 1 biological condition. For the exosome protein-enriched sample, the analysis only considered proteins present in at least 3 of 5 patients for each biological condition. Junction plakoglobin (JUP) and S100-A9 (proteins in red font) were confirmed by specific monoclonal antibodies in immunoblot analysis. Plasma-derived exosomes were obtained from 10 patients (20 samples) corresponding to the 4 different subgroups (5 patients each) and subjected to 12% polyacrylamide gel (SDS-PAGE) electrophoresis. Representative immunoblots of both proteins at the different disease times are depicted. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDL, high-density lipoprotein.
To select the proteins relevant to disease progression, we focused on the proteins that remained differentially present in the Prog-ddp subgroup. To this aim, we performed a comparative analysis between Ind-4years, Prog-dt, and Prog-ddp. The PatternLab for Proteomics software Venn diagram module pinpointed proteins uniquely identified in each subgroup. Considering proteins present in at least 3 patients per class, only 34 proteins were shared among the 3 subgroups; 2 were only found in the Ind-4years patients, 7 in Prog-dt, and 10 in Prog-ddp (Figure 2B; supplemental Data 3). Among the 10 proteins detected exclusively in the Prog-ddp subgroup, the 2 most represented (4 of 5 patients) were S100 calcium binding protein A9 (S100-A9) and JUP (Figure 2B). To confirm these results, we performed immunoblot analysis in the CLL exosomes of the 10 patients at the different disease points included in the proteomic analysis. Our results confirmed the presence of these 2 proteins in 5 of 5 patients for S100-A9 and 4 of 5 patients for JUP, whereas none or very low presence of these proteins was detected in the samples before progression or in indolent CLLs. A representative case for each protein is depicted in Figure 2C. Interestingly, both proteins are involved in cancer progression. S100-A9 is an activator of the NF-κB pathway25 and has been associated with inflammatory networks,25 and JUP is implicated in the activation of Wnt pathway.26,27
Plasma-derived exosomes with increased levels of S100-A9 are found in patients with progressive disease after disease progression and released by CLL cells
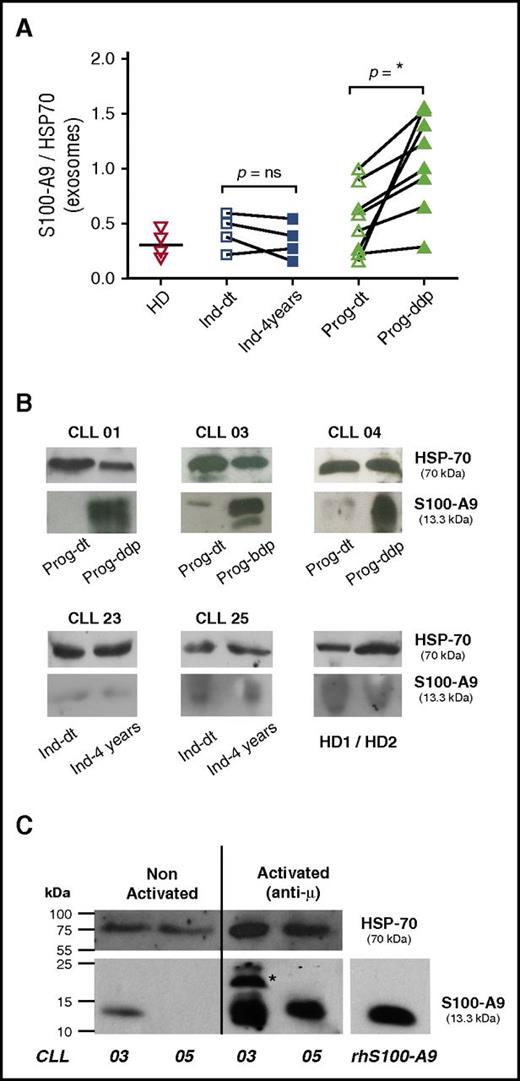
S100-A9 and the complex S100-A8/A9 are key factors leading to inflammatory cell recruitment, tumor growth, and metastasis.4,28 Our proteomic and immunoblotting analysis identified S100-A9 as 1 of the proteins specifically present in the plasma exosomes of the Prog-ddp subgroup. To confirm these results, we performed immunoblot analysis in the validation cohort (Table 1). As depicted in Figure 3A, 7 of the 8 samples showed a significant increase of S100-A9 cargo in CLL exosomes at the time of disease progression (mean, 1.12 and 0.51, respectively; Mann-Whitney test, P = .010; n = 8). In contrast, no significant differences were found within the indolent group, (Ind-dt: mean, 0.44; Ind-4years: mean, 0.33; Mann-Whitney test, P = .657; n = 4). In turn, S100-A9 levels from HDs (mean, 0.31) seemed similar to those observed in indolent and Prog-dt CLL samples. Representative cases of CLL patients from the different subgroups at both evolution times and from HDs are depicted in Figure 3B. To evaluate whether CLL cells are able to release S100-A9+ exosomes, we performed in vitro assays with CD19+ cells from patients with progressive disease during disease progression. Our results showed that after anti-IgM incubation, CLL cells are able to release exosomes carrying S100-A9, as evidenced by immunoblot analysis (Figure 3C). Overall, these findings validate proteomic analysis concerning the specific presence of S100-A9 during disease progression and suggest that these exosomes originate at least in part from CLL cells.
CLL plasma-derived exosomes show increased levels of S100-A9 protein and are released by CLL cells. (A-B) S100-A9 quantification from plasma-derived exosomes between the different subgroups; immunoblot analysis was performed after exosome protein separation. A total of 50 μg of exosome protein from the different samples was transferred onto nitrocellulose and developed with specific anti–S100-A9 monoclonal antibody. HSP-70 protein was visualized as internal charge control. Values of S100-A9 were normalized to those of HSP-70 accordingly. Results showed that S100-A9 protein was significantly higher in the plasma exosomes at the time of disease progression, whereas no or low presence of this protein was found in the exosomes of the Prog-dt subgroup, the indolent group, or the matched HDs. *P < .05. (C) Release of S100-A9+ exosomes by CLL cells; in vitro analysis was performed with CD19+ cells (100 exp10 lymphocytes incubated O.N. with anti-IgM 15 μg/ml). AIM V medium 10 mL was collected and exosomes purified as previously described in Material and Methods. Results showed that CLL cells release exosomes carrying S100-A9 protein to the culture medium after incubation with anti-IgM. *S100-A9 homodimer. Immunoblot analysis was performed as described. ns, not significant.
CLL plasma-derived exosomes show increased levels of S100-A9 protein and are released by CLL cells. (A-B) S100-A9 quantification from plasma-derived exosomes between the different subgroups; immunoblot analysis was performed after exosome protein separation. A total of 50 μg of exosome protein from the different samples was transferred onto nitrocellulose and developed with specific anti–S100-A9 monoclonal antibody. HSP-70 protein was visualized as internal charge control. Values of S100-A9 were normalized to those of HSP-70 accordingly. Results showed that S100-A9 protein was significantly higher in the plasma exosomes at the time of disease progression, whereas no or low presence of this protein was found in the exosomes of the Prog-dt subgroup, the indolent group, or the matched HDs. *P < .05. (C) Release of S100-A9+ exosomes by CLL cells; in vitro analysis was performed with CD19+ cells (100 exp10 lymphocytes incubated O.N. with anti-IgM 15 μg/ml). AIM V medium 10 mL was collected and exosomes purified as previously described in Material and Methods. Results showed that CLL cells release exosomes carrying S100-A9 protein to the culture medium after incubation with anti-IgM. *S100-A9 homodimer. Immunoblot analysis was performed as described. ns, not significant.
S100-A9 receptor EMMPRIN is expressed in leukemic cells and binds S100-A9 in vitro
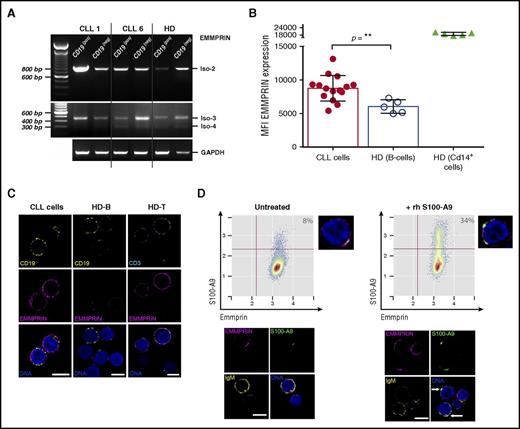
EMMPRIN is a multifunctional transmembrane protein that regulates cell proliferation and migration and promotes cancer progression.29 Because it has been shown that S100-A9 is able to bind EMMPRIN, thus mediating inflammation and cancer proliferation,28 we investigated the expression levels of this protein and its ability to bind S100-A9 protein in CLL cells. Our results showed that EMMPRIN messenger RNA (mRNA) isoform 2 but not isoform 1 was expressed in the leukemic clone, whereas low mRNA levels of isoforms 3 and 4 were found (Figure 4A). Concerning EMMPRIN protein levels, flow cytometry analysis showed a significantly higher expression in CLL cells compared with normal B cells (Figure 4B). In contrast, no significant differences were found after comparison of patients with progressive and indolent disease (data not shown). EMMPRIN expression was also characterized in T cells and macrophage/monocyte cells from HDs and patients with CLL. Different expression levels are shown in supplemental Figure 1. Confocal microscopy analysis confirmed the higher expression of EMMPRIN in CLL cells compared with those of HDs (Figure 4C). To elucidate whether EMMPRIN receptor is able to bind S100-A9 in CLL cells, we performed in vitro analysis, incubating PBMCs from progressive CLLs with rhS100-A9. Our results showed a percentage of leukemic cells expressing EMMPRIN that was able to bind rhS100-A9 (Figure 4D). These results were confirmed by confocal microscopy showing colocalization of rhS100-A9 with EMMPRIN receptor (Figure 4D, arrows).
EMMPRIN expression in CLL cells and S100-A9 binding in vitro. (A) Reverse-transcription polymerase chain reaction analysis of EMMPRIN mRNA isoforms in CLL. EMMPRIN mRNA isoforms 1 to 4 expression was evaluated in MACS-sorted B lymphocytes (CD19+) and non-B cells (CD19−) as described by Oppezzo et al.18 EMMPRIN isoform 2 is expressed in leukemic B cells at higher levels than in normal B cells, whereas isoforms 3 and 4 are also expressed but at basal levels. (B) Flow cytometry analysis of EMMPRIN protein levels in B lymphocytes. Median fluorescence intensity analysis of EMMPRIN staining in CLL and HD B cells reveals higher levels of EMMPRIN staining in CLL B cells (mean, 8.751; n = 15) compared with normal B cells (mean, 5.964; n = 5; Mann-Whitney test, P = .0037). MFI, mean fluorescence intensity. (C) EMMPRIN in situ immunostaining. Direct in situ immunolocalization of EMMPRIN (magenta) shows higher staining intensities in CLL B cells compared with their normal counterparts (CD19+ in yellow). EMMPRIN staining in normal T cells (CD3+ in cyan) was assessed as positive control. Single confocal planes are shown; deconvolution was performed with Huygens Essential 4.5 (Scientific Volume Imaging, Hilversum, The Netherlands; http://svi.nl). Scale bar, 5 µm. (D) rhS100-A9 B-cell CLL in vitro binding assay. The binding ability of rhS100-A9 (green) on B-cell CLL cells (IgM+ in yellow) expressing EMMPRIN (magenta) was evaluated on incubation of 106 cells with 5 µg/mL for 1 hour at 37°C. An increase from basal 8% to 34% of S100-A9+ cells was found. Flow cytometry analysis was performed with R/Bioconductor. Confocal microscopy analysis showed an increase in S100-A9 immunostaining and suggested colocalization of rhS100-A9 with EMMPRIN receptor, shown in white in the overlaid images (arrows). Deconvolved single confocal planes are shown. Scale bar, 5 µm.
EMMPRIN expression in CLL cells and S100-A9 binding in vitro. (A) Reverse-transcription polymerase chain reaction analysis of EMMPRIN mRNA isoforms in CLL. EMMPRIN mRNA isoforms 1 to 4 expression was evaluated in MACS-sorted B lymphocytes (CD19+) and non-B cells (CD19−) as described by Oppezzo et al.18 EMMPRIN isoform 2 is expressed in leukemic B cells at higher levels than in normal B cells, whereas isoforms 3 and 4 are also expressed but at basal levels. (B) Flow cytometry analysis of EMMPRIN protein levels in B lymphocytes. Median fluorescence intensity analysis of EMMPRIN staining in CLL and HD B cells reveals higher levels of EMMPRIN staining in CLL B cells (mean, 8.751; n = 15) compared with normal B cells (mean, 5.964; n = 5; Mann-Whitney test, P = .0037). MFI, mean fluorescence intensity. (C) EMMPRIN in situ immunostaining. Direct in situ immunolocalization of EMMPRIN (magenta) shows higher staining intensities in CLL B cells compared with their normal counterparts (CD19+ in yellow). EMMPRIN staining in normal T cells (CD3+ in cyan) was assessed as positive control. Single confocal planes are shown; deconvolution was performed with Huygens Essential 4.5 (Scientific Volume Imaging, Hilversum, The Netherlands; http://svi.nl). Scale bar, 5 µm. (D) rhS100-A9 B-cell CLL in vitro binding assay. The binding ability of rhS100-A9 (green) on B-cell CLL cells (IgM+ in yellow) expressing EMMPRIN (magenta) was evaluated on incubation of 106 cells with 5 µg/mL for 1 hour at 37°C. An increase from basal 8% to 34% of S100-A9+ cells was found. Flow cytometry analysis was performed with R/Bioconductor. Confocal microscopy analysis showed an increase in S100-A9 immunostaining and suggested colocalization of rhS100-A9 with EMMPRIN receptor, shown in white in the overlaid images (arrows). Deconvolved single confocal planes are shown. Scale bar, 5 µm.
S100-A9 is overexpressed in CLL cells, increases during disease progression, and correlates with NF-κB pathway activation
On the basis of our previous results, we evaluated S100-A9 expression at mRNA and protein levels in the leukemic cells from patients with indolent disease and from the Prog-ddp subgroup. Analysis of S100-A9 mRNA expression demonstrated that gene transcription is increased in CLL cells from patients with progressive disease (supplemental Figure 2A). At the protein level, our results showed a significant increase in the percentage of cells expressing S100-A9 in samples from patients with progressive disease when compared with those from patients with indolent disease or CD19+ cells from HDs (Figure 5A). As found with exosomes, there was an increased expression of S100-A9 at the time of progression in Prog-ddp samples compared with the same samples at disease onset (Figure 5B). These results were confirmed by confocal microscopy of PBMCs from the different subgroups (Figure 5C). Representative histograms with different S100-A9 protein expression levels in the different subgroups and in a HD are depicted in supplemental Figure 2. Supporting the hypothesis that S100-A9 is overexpressed in activated CLL cells from patients with progressive disease, we found an increased percentage of S100-A9+ B cells in lymph nodes (LNs) compared with their tumoral counterparts in the PB of the same patient (supplemental Figure 3). Interestingly, CLL cells from LNs expressing S100-A9 also showed an activated molecular profile, indicated by a higher expression of Ki-67 and survivin proteins when compared with S100-A9− CLL cells (supplemental Figure 4).
Quantification of S100-A9 expression in the leukemic clone. Flow cytometry analysis of PBMCs was performed evaluating IgM, CD5, and S100-A9+ cells between HDs and the different CLL subgroups. CD19+ cells of age-matched HDs were used to obtain reference values, and control isotypes were used for IgM and S100-A9 antibodies. (A) S100-A9 expression in indolent and progressive CLL samples during disease progression. Flow cytometry analysis was performed with specific antibodies, as described in supplemental Table 1. Five age-matched HDs to determine the physiological levels of S100-A9 in B lymphocytes and a total of 16 indolent CLLs and 18 progressive CLLs at the time of progression were evaluated. A significant increase of S100-A9 levels was found in the samples obtained from Prog-ddp CLLs (mean, 32.44) compared with samples from indolent CLLs (mean, 14.98; 1-way analysis of variance [ANOVA], P < .01). No significant changes were observed between HD samples compared with indolent CLLs (mean, 5.55 and 14.98, respectively). ns, not significant. (B) S100-A9 protein expression levels in B lymphocytes at different times of disease evolution. Five HDs, 8 Ind-4years, and 8 progressive CLLs at the time of onset and 8 during disease progression were evaluated. A significantly higher level of S100-A9 protein in the leukemic cells from the Prog-ddp samples (mean, 42.50) compared with Prog-dt (mean, 23.24) and with Ind-4years (mean, 14.64) was demonstrated (1-way ANOVA, P < .01). No significant differences were visualized comparing the expression of S100-A9 in B lymphocytes (mean, 1.38) with Ind-4years (mean, 14.64) or Prog-dt (mean, 23.24). Microscopy analysis was performed with specific anti-IgM, anti-CD5, and anti–S100-A9 (supplemental Table 1). (C) S100-A9 expression in the leukemic clone was confirmed as a discrete cytoplasmic pattern shown in red color. DNA staining was performed with methyl green as described by Oppezzo et al.22 Gray and white asterisks indicate low and high S100-A9 expression levels, respectively. Scale bar, 5μm (lower right). (D) NF-κB pathway is more activated in the samples of progressive CLLs during disease progression. Characterization of NF-κB pathway activation by immunoblot assays evaluating IκB-α and inhibitory IκB-α phosphorylation in PBMCs of the different subgroups (Prog-dt and Prog-ddp). Representative cases (10 of 20 analyzed patients) as depicted after immunoblot reaction. (E) Increased NF-κB activation in the Prog-ddp subgroup was evidenced after quantification of p65 nucleus/cytoplasm stain ratio from a total of 100 CLL cells (IgM+/CD5+) in 5 samples from each subgroup (Prog-dt and Prog-ddp; n = 10). Prog-dt samples display a low percentage of cells with the transcription factor p65 in the nucleus (mean, 30) compared with Prog-ddp samples of the same patient (mean, 50; Mann-Whitney test, P = .019). Immunofluorescent staining was performed with specific human anti-p65 transcription factor, specific anti-IgM, and anti-CD5 antibodies. (F) Positive correlation of S100-A9 expression with IκB-α phosphorylation in patients with CLL. The relationship between S100-A9 protein expression and IκB-α phosphorylation was investigated. A significant positive correlation was detected (r2 = 0.35; Spearman’s rank test, P < .001).
Quantification of S100-A9 expression in the leukemic clone. Flow cytometry analysis of PBMCs was performed evaluating IgM, CD5, and S100-A9+ cells between HDs and the different CLL subgroups. CD19+ cells of age-matched HDs were used to obtain reference values, and control isotypes were used for IgM and S100-A9 antibodies. (A) S100-A9 expression in indolent and progressive CLL samples during disease progression. Flow cytometry analysis was performed with specific antibodies, as described in supplemental Table 1. Five age-matched HDs to determine the physiological levels of S100-A9 in B lymphocytes and a total of 16 indolent CLLs and 18 progressive CLLs at the time of progression were evaluated. A significant increase of S100-A9 levels was found in the samples obtained from Prog-ddp CLLs (mean, 32.44) compared with samples from indolent CLLs (mean, 14.98; 1-way analysis of variance [ANOVA], P < .01). No significant changes were observed between HD samples compared with indolent CLLs (mean, 5.55 and 14.98, respectively). ns, not significant. (B) S100-A9 protein expression levels in B lymphocytes at different times of disease evolution. Five HDs, 8 Ind-4years, and 8 progressive CLLs at the time of onset and 8 during disease progression were evaluated. A significantly higher level of S100-A9 protein in the leukemic cells from the Prog-ddp samples (mean, 42.50) compared with Prog-dt (mean, 23.24) and with Ind-4years (mean, 14.64) was demonstrated (1-way ANOVA, P < .01). No significant differences were visualized comparing the expression of S100-A9 in B lymphocytes (mean, 1.38) with Ind-4years (mean, 14.64) or Prog-dt (mean, 23.24). Microscopy analysis was performed with specific anti-IgM, anti-CD5, and anti–S100-A9 (supplemental Table 1). (C) S100-A9 expression in the leukemic clone was confirmed as a discrete cytoplasmic pattern shown in red color. DNA staining was performed with methyl green as described by Oppezzo et al.22 Gray and white asterisks indicate low and high S100-A9 expression levels, respectively. Scale bar, 5μm (lower right). (D) NF-κB pathway is more activated in the samples of progressive CLLs during disease progression. Characterization of NF-κB pathway activation by immunoblot assays evaluating IκB-α and inhibitory IκB-α phosphorylation in PBMCs of the different subgroups (Prog-dt and Prog-ddp). Representative cases (10 of 20 analyzed patients) as depicted after immunoblot reaction. (E) Increased NF-κB activation in the Prog-ddp subgroup was evidenced after quantification of p65 nucleus/cytoplasm stain ratio from a total of 100 CLL cells (IgM+/CD5+) in 5 samples from each subgroup (Prog-dt and Prog-ddp; n = 10). Prog-dt samples display a low percentage of cells with the transcription factor p65 in the nucleus (mean, 30) compared with Prog-ddp samples of the same patient (mean, 50; Mann-Whitney test, P = .019). Immunofluorescent staining was performed with specific human anti-p65 transcription factor, specific anti-IgM, and anti-CD5 antibodies. (F) Positive correlation of S100-A9 expression with IκB-α phosphorylation in patients with CLL. The relationship between S100-A9 protein expression and IκB-α phosphorylation was investigated. A significant positive correlation was detected (r2 = 0.35; Spearman’s rank test, P < .001).
Because S100-A9 has been reported to increase NF-κB activity,26,28 we evaluated the activation of the NF-κB pathway in leukemic cells with increased expression of S100-A9. To this aim, we analyzed the Ser32/36 phosphorylation of IκB-α and the nuclear/cytoplasmic rate of the transcription factor p65 between Prog-dt and Prog-ddp samples. As shown in Figure 5D, differences in IκB-α phosphorylation at different disease points were found, indicating the release of NF-κB from inhibitory IκB-α during disease progression. Moreover, a comparison of p65 localization between Prog-dt and Prog-ddp samples of the same patient demonstrated a significant increase of this transcription factor in the nucleus of the Prog-ddp sample (Figure 5E; supplemental Figure 1D). Finally, a significant and positive correlation was found between IκB-α phosphorylation and S100-A9 expression, which suggests a link between this protein and NF-κB activity. Together, these results show that leukemic cells derived from patients with progressive disease express high levels of S100-A9 when compared with indolent CLLs and that this expression is correlated with canonical NF-κB pathway activation.
Plasma exosomes with S100-A9 cargo promote NF-κB pathway activation in leukemic cells
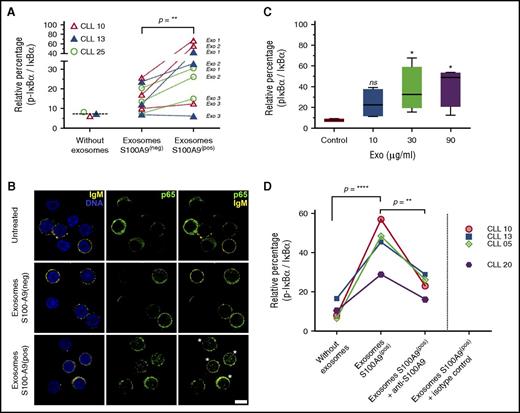
S100-A9 is able to activate the NFκB pathway in carcinoma and melanoma cells,26,30 but little evidence exists regarding its role in CLL. Taking our results into account, we hypothesized that specific expression of S100-A9 in the leukemic clone during disease progression as well as its significant increase in plasma exosomes could be one of the mechanisms triggering NF-κB activation during CLL progression. To evaluate the role of plasma exosomes with S100-A9 cargo regarding the NF-κB pathway, we obtained plasma exosomes from 3 patients with progressive disease (01, 02, and 04) at disease onset (S100-A9−) and during disease progression (S100-A9+). We incubated PBMCs of 2 patients with no mutation and 1 patient with mutation at time of disease onset and evaluated IκB-α phosphorylation and p65 nuclear translocation as regards activation of the canonical NF-κB pathway and the inducible processing of p100/p52 proteins as regards activation of the noncanonical pathway. Our results showed a clear activation of the canonical NF-κB pathway in at least 2 of the 3 plasma exosomes carrying S100-A9 in the 3 tested patients. As depicted in Figure 6A, a significant increase of IκB-α phosphorylation was observed after incubation of the exosomes extracted from the Prog-ddp subgroup (S100-A9+) but not with plasma-derived exosomes extracted from the Prog-dt subgroup (S100-A9−). The canonical NF-κB pathway activation was also confirmed by nuclear localization of p65 in the leukemic cells (Figure 6B). Interestingly, this activation seemed to be specific to the canonical NF-κB pathway but not to the noncanonical pathway (p100/p52 immunoblot analysis in supplemental Figure 5).
Plasma-derived exosomes with S100-A9 cargo promote NF-κB pathway activation in leukemic cells. (A-B) NF-κB pathway is activated after in vitro incubation with plasma-derived exosomes (Exo) carrying S100-A9. PBMCs of unmutated and mutated CLLs were incubated in RPMI 1640 and fetal bovine serum with plasma-derived exosomes (S100-A9+) extracted from Prog-ddp and plasma-derived exosomes without S100-A9 extracted from Prog-dt samples (90 minutes; 60 μg/ml of total protein). Negative and positive exosomes were categorized by immunoblot. NF-κB activation was determined as the relative percentage by densitometry of the corresponding signal from phosphorylated IκB-α/total IκB-α. “Without exosomes” indicates basal phosphorylation of IκB-α in the PBMCs of CLLs. Significant changes were observed in the rate of IκB-α phosphorylation after incubation with S100-A9+ plasma-derived exosomes compared with their counterparts (S100-A9− plasma-derived exosomes; Wilcoxon signed rank test, P = .0078). In this experiment, PBMCs at disease onset from 3 different patients, 2 with unmutated and 1 with mutated CLL (CLLs 10 and 13 and CLL 25, respectively) were incubated with plasma-derived exosomes from 3 different CLLs (Exo1, CLL 02; Exo2, CLL 05; Exo3, CLL 07) extracted at disease onset (S100-A9−) and during disease progression (S100-A9+). NF-κB pathway activation after incubation with plasma-derived exosomes carrying S100-A9 was also evidenced by p65 nuclear translocation. After 2 hours of incubation with plasma-derived exosomes (with or without S100-A9), PBMCs of the 3 CLLs were collected and stained with anti-IgM, anti-CD5, and anti-p65 transcription factor. A representative figure of the 3 patients with CLL studied is provided in Figure 5B. Green color indicates p65, and yellow staining corresponds to IgM+ cells. DNA staining with methyl green was performed. More than 95% of IgM cells for CLL 10 and 13 and 72% for CLL 25 were positive for CD5 marker (estimated by flow cytometry analysis and confocal microscopy; data not shown). Leukemic cells after incubation with exosomes carrying S100-A9 with increased nuclear localization of p65 transcription factor are highlighted by white asterisks. Scale bar, 5μm (lower right). (C) Phosphorylation of IκB-α in PBMCs of CLL after 90-minute incubation with increasing concentrations of plasma-derived exosomes carrying S100-A9. Data are reported as relative changes of phosphorylated IκB-α/total IκB-α. (D) Monoclonal antibody against S100-A9 was used to block the interaction of S100-A9+ exosomes with leukemic cells. To confirm the specificity of NF-κB activation, 60 μg of CLL plasma-derived exosomes (S100-A9+) were extracted from patient 01 and preincubated for 60 minutes with polyclonal antibody anti–S100-A9 at 6°C or with isotype control. Next, PBMCs from 4 different patients were incubated for 90 minutes at 37°C with these cocktails (S100-A9+ exosomes plus anti–S100-A9 antibody, S100-A9+ exosomes, and S100-A9+ exosomes plus isotype control antibody). Low levels of IκB-α phosphorylation were found after PBMC incubation with anti–S100-A9 (data not shown). This unspecific activation was subtracted to obtain the values corresponding to the “exosomes S100-A9+ plus anti–S100-A9 group. Our results showed a significant reduction in the 4 samples in the ratio p-IκB-α / IκB-α after CLL PBMCs were incubated with S100-A9+ exosomes plus anti–S100-A9 antibody (mean, 22) compared with CLL PBMCs incubated only with S100-A9+ exosomes (mean, 44.95; 1-way analysis of variance P < .01). Significant differences were found between CLL PBMCs incubated only with S100-A9+ exosomes compared with untreated CLLs (mean, 10.40; without exosomes).
Plasma-derived exosomes with S100-A9 cargo promote NF-κB pathway activation in leukemic cells. (A-B) NF-κB pathway is activated after in vitro incubation with plasma-derived exosomes (Exo) carrying S100-A9. PBMCs of unmutated and mutated CLLs were incubated in RPMI 1640 and fetal bovine serum with plasma-derived exosomes (S100-A9+) extracted from Prog-ddp and plasma-derived exosomes without S100-A9 extracted from Prog-dt samples (90 minutes; 60 μg/ml of total protein). Negative and positive exosomes were categorized by immunoblot. NF-κB activation was determined as the relative percentage by densitometry of the corresponding signal from phosphorylated IκB-α/total IκB-α. “Without exosomes” indicates basal phosphorylation of IκB-α in the PBMCs of CLLs. Significant changes were observed in the rate of IκB-α phosphorylation after incubation with S100-A9+ plasma-derived exosomes compared with their counterparts (S100-A9− plasma-derived exosomes; Wilcoxon signed rank test, P = .0078). In this experiment, PBMCs at disease onset from 3 different patients, 2 with unmutated and 1 with mutated CLL (CLLs 10 and 13 and CLL 25, respectively) were incubated with plasma-derived exosomes from 3 different CLLs (Exo1, CLL 02; Exo2, CLL 05; Exo3, CLL 07) extracted at disease onset (S100-A9−) and during disease progression (S100-A9+). NF-κB pathway activation after incubation with plasma-derived exosomes carrying S100-A9 was also evidenced by p65 nuclear translocation. After 2 hours of incubation with plasma-derived exosomes (with or without S100-A9), PBMCs of the 3 CLLs were collected and stained with anti-IgM, anti-CD5, and anti-p65 transcription factor. A representative figure of the 3 patients with CLL studied is provided in Figure 5B. Green color indicates p65, and yellow staining corresponds to IgM+ cells. DNA staining with methyl green was performed. More than 95% of IgM cells for CLL 10 and 13 and 72% for CLL 25 were positive for CD5 marker (estimated by flow cytometry analysis and confocal microscopy; data not shown). Leukemic cells after incubation with exosomes carrying S100-A9 with increased nuclear localization of p65 transcription factor are highlighted by white asterisks. Scale bar, 5μm (lower right). (C) Phosphorylation of IκB-α in PBMCs of CLL after 90-minute incubation with increasing concentrations of plasma-derived exosomes carrying S100-A9. Data are reported as relative changes of phosphorylated IκB-α/total IκB-α. (D) Monoclonal antibody against S100-A9 was used to block the interaction of S100-A9+ exosomes with leukemic cells. To confirm the specificity of NF-κB activation, 60 μg of CLL plasma-derived exosomes (S100-A9+) were extracted from patient 01 and preincubated for 60 minutes with polyclonal antibody anti–S100-A9 at 6°C or with isotype control. Next, PBMCs from 4 different patients were incubated for 90 minutes at 37°C with these cocktails (S100-A9+ exosomes plus anti–S100-A9 antibody, S100-A9+ exosomes, and S100-A9+ exosomes plus isotype control antibody). Low levels of IκB-α phosphorylation were found after PBMC incubation with anti–S100-A9 (data not shown). This unspecific activation was subtracted to obtain the values corresponding to the “exosomes S100-A9+ plus anti–S100-A9 group. Our results showed a significant reduction in the 4 samples in the ratio p-IκB-α / IκB-α after CLL PBMCs were incubated with S100-A9+ exosomes plus anti–S100-A9 antibody (mean, 22) compared with CLL PBMCs incubated only with S100-A9+ exosomes (mean, 44.95; 1-way analysis of variance P < .01). Significant differences were found between CLL PBMCs incubated only with S100-A9+ exosomes compared with untreated CLLs (mean, 10.40; without exosomes).
To better understand the molecular mechanism of this activation, we also performed in vitro analysis, treating CLL PBMCs at the time of disease progression with different amounts of plasma-derived exosomes carrying S100-A9. Our results suggest that phosphorylation of IκB-α is dependent on S100-A9 plasma exosome dose (Figure 6C). Finally, we preincubated these exosomes with anti–S100-A9 antibody to demonstrate the specificity of NF-κB activation in the leukemic clone. Immunoblot assays showed a decrease of IκB-α phosphorylation after incubation of PBMCs with S100-A9+ exosomes that were previously incubated with anti–S100-A9 antibody, which suggests that the interaction between cells and exosomes is blocked by specific anti–S100-A9 antibody. In contrast, high levels of IκB-α phosphorylation were maintained after incubation of plasma exosomes carrying S100-A9 or with these exosomes previously incubated with an irrelevant isotype control (Figure 6D). Together these results confirm that plasma exosomes carrying S100-A9 protein are able to promote activation of the NF-κB pathway, suggesting a specific role for these exosomes and this protein in CLL progression.
Discussion
The dynamic interplay between the tumor and its microenvironment orchestrates critical events that contribute to tumor progression and drug resistance.31 Microenvironment signals are provided not only by cell-to-cell interactions but also by different molecules such as soluble factors (cytokines, chemokines) or EVs, which play a key role in tumor-host crosstalk.32 Specifically, the functional capacity of exosomes is altered as tumors progress to an aggressive phenotype.33 We hereby report that plasma-derived exosomes of progressive and indolent CLLs throughout disease evolution display different proteomic profiles. Particularly during disease progression, we found different PPI networks associated with inflammation, tumor progression, leukemic cell infiltration, and cell survival. The PPI network AP-1 transcription factor is activated by inflammation and oxidative stress.34 In CLL, AP-1 family members have been described as being associated with NF-κB induction after CD40L activation.35 The other major network highlighted during disease progression is the pathway of class I PI3K signaling events mediated by AKT. This network is associated not only with PI3K/AKT pathway activation but also with canonical NF-κB pathway activation. The importance of synergism between NF-κB and PI3K in CLL36 and other lymphoid neoplasms37 has been extensively demonstrated.
To identify the most relevant proteins present in CLL plasma exosomes from patients with progressive disease during disease progression, we focused on the proteins that constantly seemed to be over-represented in progressive samples analyzed by liquid chromatography–tandem mass spectrometry. Among the 10 proteins exclusively present in this subgroup, we focused on S100-A9 protein, which was also found in exosomes secreted by an unmutated CLL, as described by Pagetti et al,12 whose study proposes that exosomes could induce an inflammatory phenotype in CLL and that the NF-κB pathway is activated in stromal cells. Taking these results into account and our proteomic characterization, we decided to investigate the role of S100-A9 during CLL progression. S100-A9 belongs to a family of 25 homologous intracellular calcium-binding proteins. Although S100-A9 is mainly found in heterodimers with S100-A8 (S100A8/A9, also known as calprotectin), it also exists as a homodimer, with its own functions.38 Calprotectin and S100-A9 have been largely associated with chronic inflammation and tumor promotion.4,39 Both molecules are responsible for NF-κB pathway activation, but different receptors are implicated in this process. Calprotectin activates NF-κB through TLR4,40,41 whereas S100-A9 does so through EMMPRIN receptor.25,28 Considering that CLL exosomes engage PPI networks associated with PI3K/NF-κB pathways during disease progression, that S100-A9 is 1 of the most representative proteins in exosomes of all progressive samples, and that we did not find S100-A8, we decided to evaluate the correlation between S100-A9 expression and NF-κB activity in the leukemic clone of the different subgroups. Our results showed that the elevated expression of S100-A9 in B cells of patients with disease progression is accompanied by increased activation of the NF-κB pathway. In addition, our results on EMMPRIN expression in CLL suggest that it could be 1 of the possible receptors for S100-A9. Our results showed that EMMPRIN isoform 2 is the main mRNA expressed in CLL cells, which agrees with similar results reported in other cancer cells.42 High expression of EMMPRIN in CLL cells and its interaction with S100-A9 might implicate a new activation axis for the leukemic clone during disease progression. In this line, we must take into account that EMMPRIN is able to induce MMP production in tumor cells.4,43 Interestingly, recent studies describe that overexpression of MMP-9 occurs in CLL and that elevated intracellular levels of (pro)MMP-9 correlate with advanced CLL stage and poor patient survival.4,44,45
Previous work has shown that S100-A9 is crucial in establishing the premetastatic niche, chemotherapy resistance, and subsequent metastasis in breast cancer25 and melanoma.4 The presence of S100-A9 in CLL plasma exosomes, their overexpression during leukemic progression, and the role of this protein in tumor development suggest a role in leukemia promotion and generate new and interesting questions in the pathogenesis of CLL. S100-A9 is required for the recruitment of myeloid-derived suppressive cells (MDSCs) and for suppression of an immune antitumor response.46 Specifically in CLL, Jitschin et al47 describe that through a still unknown mechanism, CLL cells are able to induce MDSCs with high expression of indoleamine-2,3-dioxygenase, which in turn becomes responsible for suppression of T-cell activation and induces suppressive regulatory T cells. Our results suggest that this unknown mechanism responsible for MDSC accumulation could originate in CLL cells expressing S100-A9 and/or exosomes carrying this protein. Another interesting question concerns the role that S100-A9 protein expressed in CLL cells could have in interacting with β2 integrins. It has been demonstrated that S100-A9 mediates neutrophil adhesion to fibronectin through activation of β2 integrins48 and that in CLL β2 integrins are essential for survival and apoptosis resistance.49 Thus, CLL cells expressing S100-A9 could interact through β2-integrins with endothelial cells to gain new survival and proliferative signals, as proposed by Maffei et al.49
Tumor-released exosomes become an efficient platform for the in vivo transfer of soluble crosstalk factors.50 The function of exosomes seems to be dependent on their protein cargoes and, in turn, dependent on the cell types from which they originate.6 Our proteomic approach demonstrates that CLL exosomes specifically contain S100-A9 during disease progression and that these exosomes activate the canonical NF-κB pathway. The fact that NF-κB activity is reduced by in vitro incubation of plasma exosomes carrying S100-A9 with specific anti–S100-A9 antibodies confirms these results. Regarding exosome function in CLL, our results support and extend the findings of Paggetti et al,12 suggesting that exosomes extracted from MEC-1 cells or, as in our case, from the plasma of patients during disease progression are able to activate the NF-κB pathway, not only in stromal cells, as described by Paggetti et al, but also in the leukemic clone. Recently, Peinado et al4 reported that melanoma-derived exosomes are able to induce the expression of S100-A8/S100-A9 in premetastatic niches after NF-κB activation, which in turn becomes activated by calprotectin and/or S100-A9.25 According to our results, an analogous loop could be envisaged in CLL when disease progression occurs. Microenvironment signals in the leukemic clone could upregulate S100-A9 expression in CLL cells, which in turn could activate the canonical NF-κB pathway, leading to an increase of proliferation and survival signaling, as suggested by the results analyzing CLL cells in LNs of patients with progressive disease, depicted in supplemental Figures 3 and 4.
In this work, we report for the first time different proteomic profiles of plasma exosomes among indolent and progressive CLLs, as well as within individual patients at the time of disease onset and during disease evolution. We describe the presence of the protein S100-A9 in CLL exosomes as well as an increased expression of this protein in leukemic cells from patients experiencing disease progression. Finally, we demonstrate that exosomes with S100-A9 cargo are able to increase NF-κB activity in CLL cells of patients with typical nonprogressive disease. This report proposes a new origin of NF-κB activation in CLL and highlights the importance of exosomes as extracellular mediators in the promotion of CLL progression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mirta Giordano and Romina Gamberale for critical comments and Hugo Naya for technical assistance in statistical analysis.
This work was supported by grants from Agencia Nacional de Investigación e Innovación (FCE_1_2011_1_7273 and FMV_2011_7323) (P.O.) and Comisión Sectorial de Investigación Científica (CSIC Proyectos I+D_2014) (P.O.).
Authorship
Contribution: D.P., N. Sotelo, and N. Seija performed experiments, collected chronic lymphocytic leukemia (CLL) samples, prepared figures, and wrote the paper; S.S. and C.A. performed experiments and collected CLL samples; R.D. and M.G. performed mass spectrometry and proteomic analysis; E.S. performed electronic microscopy analysis; V.I., C.O., A.I.L., R.G., and G.D. performed clinical activities and data collection for patients with CLL; and P.O. designed research, coordinated the study and data analysis, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pablo Oppezzo, Institut Pasteur de Montevideo, Research Laboratory on Chronic Lymphocytic Leukemia, Mataojo 2020, Montevideo (11400), Uruguay; e-mail: poppezzo@pasteur.edu.uy.
References
Author notes
D.P., N. Sotelo, and N. Seija contributed equally to this work.


![Figure 5. Quantification of S100-A9 expression in the leukemic clone. Flow cytometry analysis of PBMCs was performed evaluating IgM, CD5, and S100-A9+ cells between HDs and the different CLL subgroups. CD19+ cells of age-matched HDs were used to obtain reference values, and control isotypes were used for IgM and S100-A9 antibodies. (A) S100-A9 expression in indolent and progressive CLL samples during disease progression. Flow cytometry analysis was performed with specific antibodies, as described in supplemental Table 1. Five age-matched HDs to determine the physiological levels of S100-A9 in B lymphocytes and a total of 16 indolent CLLs and 18 progressive CLLs at the time of progression were evaluated. A significant increase of S100-A9 levels was found in the samples obtained from Prog-ddp CLLs (mean, 32.44) compared with samples from indolent CLLs (mean, 14.98; 1-way analysis of variance [ANOVA], P < .01). No significant changes were observed between HD samples compared with indolent CLLs (mean, 5.55 and 14.98, respectively). ns, not significant. (B) S100-A9 protein expression levels in B lymphocytes at different times of disease evolution. Five HDs, 8 Ind-4years, and 8 progressive CLLs at the time of onset and 8 during disease progression were evaluated. A significantly higher level of S100-A9 protein in the leukemic cells from the Prog-ddp samples (mean, 42.50) compared with Prog-dt (mean, 23.24) and with Ind-4years (mean, 14.64) was demonstrated (1-way ANOVA, P < .01). No significant differences were visualized comparing the expression of S100-A9 in B lymphocytes (mean, 1.38) with Ind-4years (mean, 14.64) or Prog-dt (mean, 23.24). Microscopy analysis was performed with specific anti-IgM, anti-CD5, and anti–S100-A9 (supplemental Table 1). (C) S100-A9 expression in the leukemic clone was confirmed as a discrete cytoplasmic pattern shown in red color. DNA staining was performed with methyl green as described by Oppezzo et al.22 Gray and white asterisks indicate low and high S100-A9 expression levels, respectively. Scale bar, 5μm (lower right). (D) NF-κB pathway is more activated in the samples of progressive CLLs during disease progression. Characterization of NF-κB pathway activation by immunoblot assays evaluating IκB-α and inhibitory IκB-α phosphorylation in PBMCs of the different subgroups (Prog-dt and Prog-ddp). Representative cases (10 of 20 analyzed patients) as depicted after immunoblot reaction. (E) Increased NF-κB activation in the Prog-ddp subgroup was evidenced after quantification of p65 nucleus/cytoplasm stain ratio from a total of 100 CLL cells (IgM+/CD5+) in 5 samples from each subgroup (Prog-dt and Prog-ddp; n = 10). Prog-dt samples display a low percentage of cells with the transcription factor p65 in the nucleus (mean, 30) compared with Prog-ddp samples of the same patient (mean, 50; Mann-Whitney test, P = .019). Immunofluorescent staining was performed with specific human anti-p65 transcription factor, specific anti-IgM, and anti-CD5 antibodies. (F) Positive correlation of S100-A9 expression with IκB-α phosphorylation in patients with CLL. The relationship between S100-A9 protein expression and IκB-α phosphorylation was investigated. A significant positive correlation was detected (r2 = 0.35; Spearman’s rank test, P < .001).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/6/10.1182_blood-2017-02-769851/4/m_blood769851f5.jpeg?Expires=1769100952&Signature=iKp3nWfzVgmM2IhwYO7g7YjGFyyUhNUveCcPrQT911Vz3uQjb2JK1KY~C--y-jjyB~KZliUULlojY0FzQHhbl7bJakOuXUwZI9j58DbQ87G2dEtY2SVcesRBaUA5WSN8A40jcGDWRvkPx9XHLmOQI1MifBmt0Ncb~8O2BKYxX1oFAUgDqa3wBzzdRoDlAjR1giAlbwIiXX3vSTnQczLsd0XMXbwprUdiPtzmQ1Fdw7-mW5mefzKKl9fzTP4cwBLlqh3Jy9mDnmE-RonJLdM~aklk999D8Wi2AcKQlq9EcqjrOfdZK~qqc4P6DIZMmz5u4xZq8A7o5A3ESqzJeovHdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal