Key Points
EVs derived from DLBCL cells can be traced from and provide insight into cell of origin.
Mutated RNAs may be preferentially packaged into EVs, and this could enable disease monitoring through liquid biopsy.
Abstract
The biological role of extracellular vesicles (EVs) in diffuse large B-cell lymphoma (DLBCL) initiation and progression remains largely unknown. We characterized EVs secreted by 5 DLBCL cell lines, a primary DLBCL tumor, and a normal control B-cell sample, optimized their purification, and analyzed their content. We found that DLBCLs secreted large quantities of CD63, Alix, TSG101, and CD81 EVs, which can be extracted using an ultracentrifugation-based method and traced by their cell of origin surface markers. We also showed that tumor-derived EVs can be exchanged between lymphoma cells, normal tonsillar cells, and HK stromal cells. We then examined the content of EVs, focusing on isolation of high-quality total RNA. We sequenced the total RNA and analyzed the nature of RNA species, including coding and noncoding RNAs. We compared whole-cell and EV-derived RNA composition in benign and malignant B cells and discovered that transcripts from EVs were involved in many critical cellular functions. Finally, we performed mutational analysis and found that mutations detected in EVs exquisitely represented mutations in the cell of origin. These results enhance our understanding and enable future studies of the role that EVs may play in the pathogenesis of DLBCL, particularly with regards to the exchange of genomic information. Current findings open a new strategy for liquid biopsy approaches in disease monitoring.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common type of non-Hodgkin lymphoma, is an aggressive and biologically heterogeneous disease. DLBCL is cured in ∼60% of patients with 6 cycles of rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone.1 Patients who are not cured with frontline therapy are typically treated with salvage chemotherapy followed by autologous stem cell transplant in those who are eligible.2 Unfortunately, patients who relapse after or are refractory to second-line therapy have markedly poor prognoses associated with aggressive disease and chemotherapy resistance.3,4 The causes of chemoresistance and clonal evolution in DLBCL are not entirely understood, but are likely rooted in genetic and epigenetic events and neoplastic alterations of its transcriptional programming.
Gene expression profiling identified 2 major molecular subtypes of DLBCL: germinal center B-cell–like (GCB) and activated B-cell–like (ABC).5 GCB DLBCLs arise within the germinal center (GC) of the secondary lymphoid follicle, the site of massive clonal expansion and somatic hypermutation of immunoglobulins that occurs after an antigen-dependent activation of B cell.6 ABC DLBCLs develop from the late GC and post-GC stage and are associated with constitutive activation of the NF-κB pathway. They are characterized by poorer response to chemotherapy than the GCB DLBCL subtype.7 The enzymatic machinery that mediates the GC reaction is error prone and can introduce point mutations and changes in DNA methylation in the genome, resulting in a physiological state of genomic and epigenetic instability leading to malignant transformation.8
DLBCL has also been categorized using oxidative phosphorylation/B-cell receptor/host response clusters and epigenetic patterns of methylation and histone modification.9,10 Increased disruption of methylation correlates with worse clinical outcomes.10 These categories are often independent of the cell of origin classification and provide further insight into a complex biology of DLBCL.
The biological significance of cell-free circulating DNA (cfDNA) and extracellular vesicles (EV) is emerging as a potential contributor to tumor pathogenesis. EVs are released from cells and can bind to or be taken up by other cells with resulting biological effects. They have been shown to modify the tumor microenvironment and contribute to progression of disease in various malignancies.11-13 Exosomes, the smallest EVs, have been studied extensively in melanoma and pancreatic cancer, in which they travel to sites of metastases and promote spread of disease.11,14 EVs can also interact with their environment by release and exchange of their protein and RNA contents. For example, mantle cell lymphoma–derived exosomes are preferentially taken up by mantle cell lymphoma cells.15 In Waldenstrom macroglobulinemia, the characteristically mutated protein MyD88L265P is transferred by EVs and results in activation of endogenous MyD88.16 Although uptake of EVs can be nonspecific, particular cellular events can precede uptake. For example, in myeloma and lymphoma cells, Bcl-xL cleavage leads to inclusion of exosomes.17 Exosomes are enriched in small RNAs, including microRNAs (miRNAs).18 Chronic lymphocytic leukemia cells have a particular miRNA signature.19 Small RNA profiles from a panel of human B cells, including 1 DLBCL cell line (BJAB) and its secreted exosomes, revealed nonrandomly distributed subsets of miRNA species with 3′ end nucleotide additions between B cells and exosomes.20
Increasing evidence supports a role for EVs in DLBCL progression and response or resistance to therapy. Transformation of a mouse embryonic cell line to DLBCL has shown that larger EVs called microvesicles from the transformed cell lines contained a protein (nonreceptor tyrosine kinase focal adhesion kinase) that promoted cell survival.21 In the DLBCL cell line OCI-Ly3, exosome-mediated transport of Wnt3a protein impacts the overall population equilibrium of cells.22 DLBCL cell line exosomal contents have also been associated with impaired sensitivity to chemotherapy agents doxorubicin and pixantrone, which is dependent on the expression of the adenosine triphosphate–transporter A3.23 Exosomes containing CD20 were able to shield target cells from anti-CD20 antibody attack, revealing a possible role of resistance to immunotherapy in DLBCL patients,24 again through expression of adenosine triphosphate–transporter A3.
Comprehensive characterization of the RNA content of DLBCL-derived EVs could enable us to gain a significant understanding of the biology of the disease. Few studies have been published in this area. We set out to learn more about secretion of EVs by lymphomas, to characterize their RNA content and compare with that of normal B-cell–derived EVs, and to harness this knowledge for therapeutic purposes.
Materials and methods
Cell culture conditions
DLBCL cell lines of the ABC subtype (HBL1, OCI-Ly3, TMD8) and GCB subtype (OCI-Ly1 and OCI-Ly7) were obtained from Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures) and were cultured following DSMZ recommendations. (For more details, see supplemental Materials and methods, available on the Blood Web site.)
Primary samples were obtained from Weill Cornell Medical Center under institutional review board approval and were cultured as cell lines above. A summary of the cell lines and primary cells samples is listed in supplemental Table 1.
EV isolation
Isolation of EVs was done after at least 48 hours of incubation in serum-free media by ultracentrifugation using the Beckman Coulter Ultracentrifuge Optima L90K, SW28 rotor. Serial centrifugation was performed, and supernatant was collected at each step. Size and concentration of EVs were analyzed using NanoSight NS500 nanoparticle characterization system (NanoSight, Malvern Instruments, Malvern, Worcestershire, United Kingdom). Protein concentration was determined by spectrophotometer NanoDrop ND-1000 (Thermo Fisher Scientific). (For more details, see supplemental Materials and methods.)
Flow cytometric analysis
EV yield was quantified using NanoDrop, and then 5 μg of EVs were incubated with 10 μL of 3.9-μm latex beads, surfactant-free aldehyde/sulfate, and 4% solids (Interfacial Dynamics) for 15 minutes at room temperature in a 1.5-mL tube. The tube was filled to 1 mL and incubated overnight at 4°C. Glycine was then added (110 μL of 1 M) and incubated at room temperature for 30 minutes. After multiple washes of 10-μL latex beads, bound EVs were incubated with 50 μL of the following: antiexosomal marker CD63-PE, CD20-PerCP, and/or CD3-FITC (BD) or CD10-FITC (BD) for 1 hour. Fluorescence-activated cell sorter (FACS) analysis was performed using MACSQuant Analyzer 10 (Miltenyi Biotec) and analyzed using the FlowJo software package (TreeStar). (For more details, see supplemental Materials and methods.)
Western blotting
EV and matching whole cell lysates from OCI-Ly1 and HBL1 cells were prepared using 50 mM Tris HCl pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA lysis buffer, and complete protease inhibitor cocktail (Roche). Protein lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes, which were probed with the primary antibodies: Alix (BD 634501, at 1:500), TSG101 (abcam 125011, at 1:500), CD81 (Santa Cruz 166029, at 1:1000), and HSP90B1 (R&D 7606, at 1:3000). (For more details, see supplemental Materials and methods.)
EV exchange
DLBCL cell lines (OCI-Ly1, OCI-Ly3) were pulse labeled with green fluorescent dye SYTO RNASelect (ThermoFisher) for 120 minutes at 37°C. Cells were cultured for 48 hours and then spun at 1200 rpm for 5 minutes. Supernatant was collected and incubated for 20 minutes with 40 μL ribonuclease to eliminate free RNA and then was passed through a 0.2-μm filter to eliminate debris and larger vesicles. Filtered supernatant from stained OCI-Ly1 cells was cultured with unstained OCI-Ly3 cells. The positive control experiment was performed in which filtered supernatant from unstained OCI-Ly1 cells was cultured with unstained OCI-Ly3 cells for 48 hours. This experiment was also conducted by coculturing isolated EVs from stained OCI-Ly1 and HBL1 cells with HK cells. FACS analysis was then performed to detect SYTO RNA (FITC channel) and CD63-PE using MACSQuant Analyzer 10 (Miltenyi Biotec) and analyzed using the FlowJo software package (FLowJo LLC).
Confocal microscopy
To trace EV uptake by recipient stromal HK cells with fluorescence microscopy, cells were seeded into 6-well plates and incubated with the green SYTO RNASelect–stained exosomes released by the lymphoma cell lines OCI-Ly1 and HBL1 for 3 hours; then, cells were fixed with 4% paraformaldehyde for 20 minutes, permeabilized with 0.1% Triton X-100, and stained with tetramethylrhodamine-phalloidin (α-actin) and 4′,6-diamidino-2-phenylindole (DAPI; nucleus). Images were obtained with a Zeiss LSM 880 using a 63× oil objective.
RNA extraction
Isolation of nucleic acids was performed with the miRCURY RNA Isolation Cell and Plant kit (Exiqon). A final RNA purification step was performed using Qiagen RNeasy Mini Kit columns. To determine integrity and purity of the isolated RNA, samples were evaluated on the 2100 Agilent Bioanalyzer electropherogram. RNA was quantified with Qubit 3.0 Fluorometer (Life Technologies).
Preparation of complementary DNA libraries
Total RNA ribo-depleted strand-specific paired-end complementary DNA libraries were prepared following TruSeq Stranded Total kit and were sequenced with 50-bp paired end approach on a HiSeq 2500 Sequencing System (Illumina). A small RNA-seq Analysis Pipeline was developed that included quality control (QC), removing Illumina TruSeq adapters, mapping the reads, summarizing the data, and assigning reads to annotated genes.
RNA sequencing analyses
Reads were trimmed for adapter sequences using Flexbar (v.2.4)25 and aligned to hg19 using STAR aligner (v.2.4.0).26 Transcript quantification was performed by RSEM (v.1-2.22)27 using transcript annotation from GENCODE (v19).28 Grouping of transcripts by transcript class was performed according to Ensembl biotypes classification (http://www.gencodegenes.org/gencode_biotypes.html). Differential gene expression (DE) analysis was performed using DESeq2 (v.1.12.4)29 using a filtering criteria of adjusted P value ≤.1 among groups to identify differentially expressed transcripts. Supplemental Table 1 summarizes the groups and samples used in the DE analysis. The DE genes are listed in supplemental Table 2. (For more details, see supplemental Materials and methods.)
Visualization of shared transcripts
Transcript intersecting sets between EVs from different cell lines were performed using UpSet plots in order to visualize overlapping transcripts.30
IPA analysis
Differentially expressed transcripts from EVs and whole cells were subjected to ingenuity pathway analysis (IPA) (Qiagen Inc) for mapping biological functions, identifying gene networks and canonical pathways that were significantly enriched for each EV/whole cell comparison (supplemental Table 3). Ingenuity Knowledge Base genes were used as a reference set, and cutoffs were logFC > |1.5| and P < .01.
Cell line mutational analysis
DNA was extracted from DLBCL cell lines for whole exome sequencing as previously described.31,32 Novel coding region single nucleotide variants were defined as those that were not present in SNP132. These single nucleotide variants were then further filtered by sequencing depth (≥20×) and variant percentage (≥25%).
EV and whole cell mutational analysis
RNA-seq reads generated from whole cells and EVs were used for single nucleotide polymorphism calling. Sequencing data preprocessing was carried out using JAX civet pipeline (https://github.com/TheJacksonLaboratory/civet, version 4.3.7) developed by Jackson Laboratories. All synonymous mutations were filtered out, followed by filtering of nonsynonymous mutations with <20% allele frequency. Mutations that passed these filters were compared between the cell of origin and corresponding EVs. The significance of overlap between EV mutations and whole cell mutations was evaluated using the hypergeometric test. (For more details, see supplemental Materials and methods.)
Results
EVs from DLBCL cells reflect the cell of origin
EVs were purified from 5 DLBCL cell lines (OCI-Ly1, OCI-Ly3, OCI-Ly7, HBL1, and TMD8), 1 primary DLBCL sample of GCB subtype, and 1 normal set of B cells from reactive human tonsils using serial ultracentrifugation from a starting amount of 80 000 to 100 000 cells in culture. Size and concentration of EVs were determined using NanoSight technology and ranged from 84 to 211 nm and 4.88 to 14.91 × 108 particles per milliliter, respectively (Figure 1A; supplemental Figure 1).
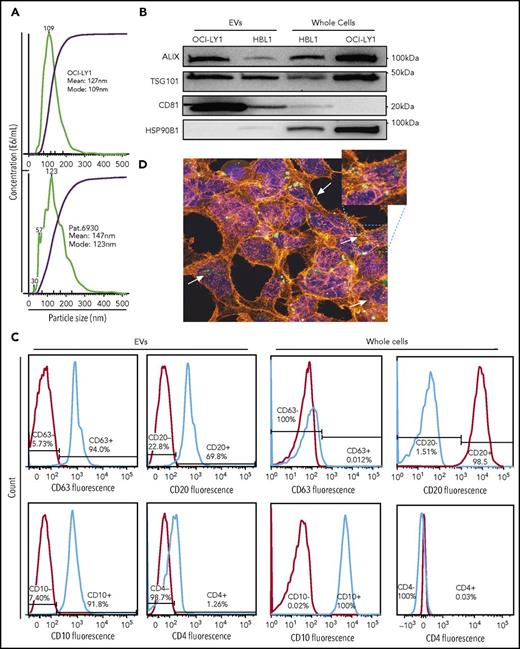
Isolation of EVs from DLBCL cell lines and primary sample. (A) Size distribution and concentration of EVs isolated from OCI-Ly1 cell line (top) and a primary DLBCL sample of GCB subtype (bottom) by ultracentrifugation determined by the NanoSight instrument. (B) Western blot for the exosomal proteins ALIX (100 kDa), TSG101 (46 kDa), CD81 (26 kDa), and as a negative control, HSP90B1 protein (90 kDa). (C) FACS analysis of exosomal surface markers demonstrates expression of CD63 in OCI-Ly1–derived EVs but not in the parental whole cells, expression of B-cell surface marker CD20, GC marker CD10, but not T-cell marker CD4 in both derived EVs and parental OCI-Ly1 cells. (D) Confocal fluorescence microscope image shows uptake of EVs released by lymphoma OCI-LY1 cells (green) by stromal HK cells (blue = DAPI; red = α-actin).
Isolation of EVs from DLBCL cell lines and primary sample. (A) Size distribution and concentration of EVs isolated from OCI-Ly1 cell line (top) and a primary DLBCL sample of GCB subtype (bottom) by ultracentrifugation determined by the NanoSight instrument. (B) Western blot for the exosomal proteins ALIX (100 kDa), TSG101 (46 kDa), CD81 (26 kDa), and as a negative control, HSP90B1 protein (90 kDa). (C) FACS analysis of exosomal surface markers demonstrates expression of CD63 in OCI-Ly1–derived EVs but not in the parental whole cells, expression of B-cell surface marker CD20, GC marker CD10, but not T-cell marker CD4 in both derived EVs and parental OCI-Ly1 cells. (D) Confocal fluorescence microscope image shows uptake of EVs released by lymphoma OCI-LY1 cells (green) by stromal HK cells (blue = DAPI; red = α-actin).
We confirmed using western blotting that the purified fraction was enriched with exosomes, by using antibodies against the exosomal markers Alix, CD81, and TSG101 (Figure 1B). Enrichment with exosomes was also confirmed by FACS staining purified EVs with anti-CD63 antibody (Figure 1C). Additional B-lymphocyte lineage markers, including CD20, CD4, and CD10, were analyzed by FACS in order to trace the cellular origin of EVs (Figure 1C). DLBCL-derived EVs were positive for the B-cell marker CD20, and negative for the T-cell marker CD4. EVs from OCI-Ly1 cell line, which is classified as GCB-derived based on gene expression profiling,33 were concordantly positive for the GC marker CD10 (Figure 1C). These findings underscore that EVs carry cell surface antigen expression corresponding to their cell of origin, allowing characterization of EV “lineage.”
Communication between DLBCL cells and primary cells can occur via uptake of EVs and their content
In order to address the potential functional role of EVs in the pathogenesis of DLBCL, we studied the ability of lymphomas to exchange EVs and their contents. We used SYTO RNASelect (SYTO RNA) dye, which labels all RNA species in the cell of origin and allows tracing the passage of these labeled RNAs after they are packaged into EVs and released. We stained OCI-Ly1 cells with SYTO RNA dye and cultured them for 48 hours. The supernatant containing EVs was cleared of cells and cell debris through filtration and then cultured with unstained OCI-Ly3 cells. Reciprocally, the filtered supernatant containing EVs from stained OCI-Ly3 cells was cultured with unstained OCI-Ly1 cells. The experiment was also performed with isolated EVs from stained OCI-Ly1 and HBL1 cells cultured with unstained primary HK stromal cells. We included unstained cells as control.
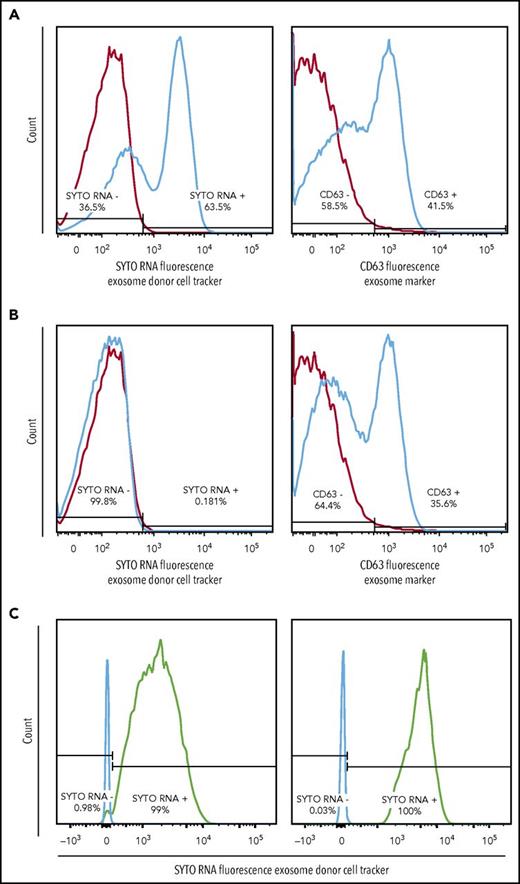
We observed uptake of OCI-Ly1 EVs by OCI-Ly3 cells and uptake of OCI-Ly1 EVs and HBL1 EVs by HK stromal cells, as unstained cells, including primary HK stromal cells, became positive for the fluorescent SYTO-labeled RNAs and the exosomal surface marker CD63 after incubation with labeled exosomes (Figure 1D and 2A). As a control, OCI-Ly3 cells were cultured with unstained OCI-Ly1 supernatant (Figure 2B), and HK cells were cultured with unstained OCI-Ly1 supernatant (not shown). We were also able to demonstrate uptake of green SYTO-labeled EVs released by OCI-Ly1 and HBL1 DLBCL cells by primary tonsillar lymphocytes using flow cytometry (Figure 2C), and uptake by stromal HK cells stained with DAPI and tetramethylrhodamine-phalloidin using confocal microscopy (Figure 1D). These findings confirm that EVs and their contents are exchanged between lymphoma cells and between lymphoma and normal cells, providing a means of sharing genetic information.
EV exchange by DLBCL cell lines and uptake by normal tonsillar cells. (A) Positive SYTO RNA fluorescent stain of supernatant from recipient OCI-Ly3 cells previously incubated with EVs from stained donor OCI-Ly1 cells (left panel) and positive stain for the general exosome marker CD63 (right panel), indicating uptake of RNAs from stained OCI-Ly1 donor cells. (B) Control negative SYTO RNA fluorescent staining of supernatant from recipient OCI-Ly3 cells incubated with EVs of unstained OCI-Ly1 donor cells (left panel), and positive staining for CD63 (right panel). (C) Positive SYTO RNA fluorescent stain in recipient normal tonsillar lymphocytes incubated with EVs isolated from previously stained donor OCI-Ly1 (left panel) or HBL1 (right panel).
EV exchange by DLBCL cell lines and uptake by normal tonsillar cells. (A) Positive SYTO RNA fluorescent stain of supernatant from recipient OCI-Ly3 cells previously incubated with EVs from stained donor OCI-Ly1 cells (left panel) and positive stain for the general exosome marker CD63 (right panel), indicating uptake of RNAs from stained OCI-Ly1 donor cells. (B) Control negative SYTO RNA fluorescent staining of supernatant from recipient OCI-Ly3 cells incubated with EVs of unstained OCI-Ly1 donor cells (left panel), and positive staining for CD63 (right panel). (C) Positive SYTO RNA fluorescent stain in recipient normal tonsillar lymphocytes incubated with EVs isolated from previously stained donor OCI-Ly1 (left panel) or HBL1 (right panel).
RNAs from EVs contain protein coding and noncoding species with important roles in B-cell development
Transcript expression was highly correlated between samples from EVs derived from cell lines (mean Pearson correlation of 0.91), as were the transcripts in EVs from primary samples (Pearson correlation of 0.96) (supplemental Figure 2A), and DLBCL cell lines (mean Pearson correlation of 0.88) (supplemental Figure 2B). In contrast, EVs derived from primary DLBCL and normal whole cell samples had poor correlation with cell-line EVs (mean correlation of 0.25) (supplemental Figure 2A). Considering that all samples passed strict QC parameters for RNA quality, library preparation, and sequencing run QC (on average we obtained >25 million reads per sample with >97% mapped reads), the differences in transcript expression between EVs derived from cell lines and primary samples point to possible biological diversity. In order to understand the differences between EVs derived from different cell types, we delineated the findings based on transcript categories.
Further analysis of RNAs revealed that EVs derived from normal B cells and from B-cell lymphomas contained a variety of RNA species, including protein-coding RNAs, long intergenic noncoding RNAs, miRNAs, small nucleolar RNAs (snoRNAs), small nuclear RNA (snRNA), and antisense RNAs (Figure 3). The most prominent RNA species in DLBCL cell lines and EVs from these cell lines were protein-coding RNAs, snoRNAs, and snRNAs (Figure 3A-B). We also found that the composition of the RNA species in EVs differed from that of parental cells (Figure 3B,D,F; supplemental Figure 3) and varied between EVs derived from normal B cells and lymphoma cells. For example, protein-coding RNAs were most abundant in whole cells and cell line–derived EVs, whereas snRNAs were most abundant in EVs from normal B cells and primary DLBCLs (Figure 3D,F). The difference may be biological. The impact of RNA from EVs in vivo may be primarily through activity of snRNAs in addition to the protein coding function. Although fewer protein coding RNAs were isolated from patient samples compared with cell lines, this RNA type was prominent in all samples analyzed. We were not able to obtain a larger quantity of fresh viable primary samples, so such observations remain preliminary and require further future studies to make definitive conclusions. The differences in RNA species packaged in EVs may be biological but may also reflect that primary samples do not grow as well in culture as cell lines.
RNA species found in EVs and cells. (A) DLBCL cell lines OCI-Ly3, HBL1, TMD8, OCI-Ly1, and OCI-Ly7, (B) EVs from DLBCL cell lines OCI-Ly3, HBL1, TMD8, OCI-Ly1, and OCI-Ly7, (C) whole cell DLBCL primary samples DLBCL1, DLBCL2, DLBCL3, DLBCL4, DLBCL5, (D) EVs from DLBCL primary sample DLBCL6, (E) whole cell normal B cells NB1, NB2, and NB3, and (F) EVs from normal B cells B2. EVs contain a variety of RNA species with enrichment for noncoding RNAs, long intergenic noncoding RNAs (lincRNAs), snoRNAs, and snRNAs. Quantities are displayed as the sum of average transcripts per million (TPM) per RNA class. rRNA, ribosomal RNA.
RNA species found in EVs and cells. (A) DLBCL cell lines OCI-Ly3, HBL1, TMD8, OCI-Ly1, and OCI-Ly7, (B) EVs from DLBCL cell lines OCI-Ly3, HBL1, TMD8, OCI-Ly1, and OCI-Ly7, (C) whole cell DLBCL primary samples DLBCL1, DLBCL2, DLBCL3, DLBCL4, DLBCL5, (D) EVs from DLBCL primary sample DLBCL6, (E) whole cell normal B cells NB1, NB2, and NB3, and (F) EVs from normal B cells B2. EVs contain a variety of RNA species with enrichment for noncoding RNAs, long intergenic noncoding RNAs (lincRNAs), snoRNAs, and snRNAs. Quantities are displayed as the sum of average transcripts per million (TPM) per RNA class. rRNA, ribosomal RNA.
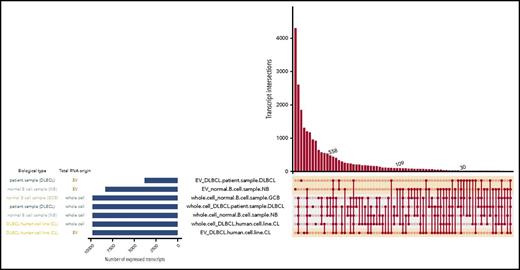
We further investigated transcripts shared between different samples. Generally, cell lines shared a higher fraction of their transcripts with each other than with their respective EVs (Figure 4). The left panel of Figure 4 displays the number of expressed transcripts for each sample type. The right panel shows shared transcripts between sample types connected and marked by circles. The number of shared transcripts is indicated in the y-axis. DLBCL cell lines and their corresponding EVs shared a total of 558 transcripts. The primary DLBCL sample shared 30 transcripts with corresponding EVs from DLBCL cell lines, and the DLBCL primary sample shared 109 transcripts. It is notable that more transcripts were shared among DLBCL EVs from cell lines and primary sample than the primary DLBCL sample shared with corresponding EVs.
Intersecting sets of transcripts across EVs and whole cell content. Depiction of all possible transcript overlap sets among the DLBCL cell lines, primary samples, normal B cells, and their corresponding EVs. Samples are listed on the left with the blue bar indicating the number of expressed RNA transcripts. On the right panel, the average of TPM values was plotted in this graph for each transcript isoform from each sample type (EV-normal B, EV-primary DLBCL, etc). Then, to restrict the analysis to the most abundant transcripts, the top 5% expressed transcripts from each sample type were selected and analyzed for overlapping transcripts. The intersections of sets are a matrix in which the rows represent the sets and the columns represent their intersections. The number of shared transcripts is indicated in the y-axis. Overlaps are sorted in decreasing size, and the shaded circles connected by solid lines in the lower panel indicate the intersecting samples.
Intersecting sets of transcripts across EVs and whole cell content. Depiction of all possible transcript overlap sets among the DLBCL cell lines, primary samples, normal B cells, and their corresponding EVs. Samples are listed on the left with the blue bar indicating the number of expressed RNA transcripts. On the right panel, the average of TPM values was plotted in this graph for each transcript isoform from each sample type (EV-normal B, EV-primary DLBCL, etc). Then, to restrict the analysis to the most abundant transcripts, the top 5% expressed transcripts from each sample type were selected and analyzed for overlapping transcripts. The intersections of sets are a matrix in which the rows represent the sets and the columns represent their intersections. The number of shared transcripts is indicated in the y-axis. Overlaps are sorted in decreasing size, and the shaded circles connected by solid lines in the lower panel indicate the intersecting samples.
Pathway analysis of the coding genes present in normal B cells (naive and GC B cells) and EVs secreted by these cells revealed an enrichment in EVs for cellular functions, such as molecular transport, cell morphology, cellular development, cellular growth and proliferation, and associated network functions, including cell-to-cell signaling and interaction (Figure 5A; DEG_Normal-EV_vs_Normal-Whole section in supplemental Table 3). As expected, EVs from ABC cell lines are enriched with molecules related with diseases as cancer, hematological disease, and immunological disease (Figure 5B; DEG_ABC-EV_vs_ABC-Whole in section supplemental Table 3). In addition, we carried out differential expression analysis between the RNAs isolated from EVs of ABC and GCB DLBCL cell lines. Pathway analysis confirmed known differences between transcriptional profiles of these 2 molecular subtypes of lymphoma.34 We found that RNA species from oxidative phosphorylation pathways differentiated ABC- and GCB-cell line–derived EVs (P = 6.62 E-09). Top canonical pathways also included mitochondrial dysfunction (P = 9.81 E-08) and EIF2 signaling (P = 2.1 E-04) (supplemental Table 3).
IPA core analysis–based associated networks. (A) normal EVs vs normal whole cells: Cell-To-Cell Signaling and Interaction, Nervous System Development and Function, Neurological Disease (score 38) and (B) ABC EVs vs ABC whole cell lines: cancer, hematological disease, immunological disease (score 30). Red represents positive enrichment, and green represents negative enrichment in the EVs compared with the respective whole cell. Links of validated genes and other genes or molecules are represented with a continuous (direct interaction) or discontinuous line (indirect interaction).
IPA core analysis–based associated networks. (A) normal EVs vs normal whole cells: Cell-To-Cell Signaling and Interaction, Nervous System Development and Function, Neurological Disease (score 38) and (B) ABC EVs vs ABC whole cell lines: cancer, hematological disease, immunological disease (score 30). Red represents positive enrichment, and green represents negative enrichment in the EVs compared with the respective whole cell. Links of validated genes and other genes or molecules are represented with a continuous (direct interaction) or discontinuous line (indirect interaction).
We determined that RNA content in EVs reflects the cell of origin and its biology, but also contains unique species. This suggests EVs may play a broad role in pathobiology of DLBCLs.
Mutational profiles of EV-derived RNAs reflect genomic landscape in the cell of origin
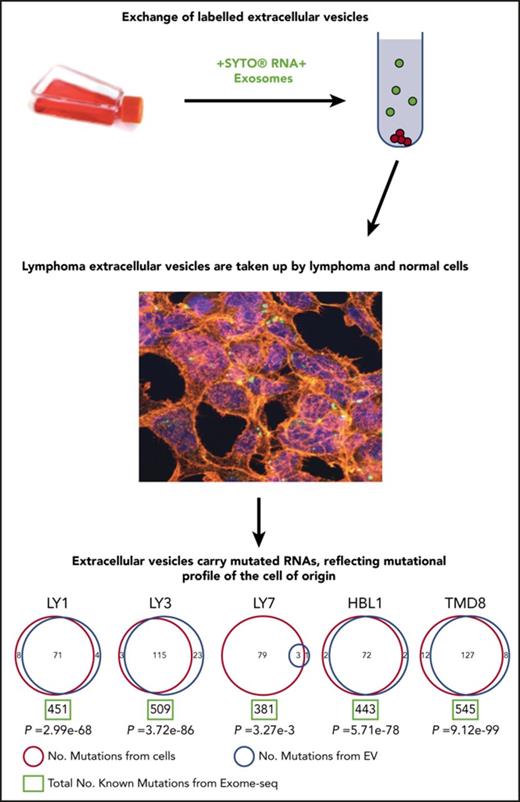
Disease monitoring using liquid biopsy and sampling cfDNA is currently a state-of-the-art approach. Because EV-derived RNA is abundant and stable, we investigated whether EVs could inform about the mutational profile in the cell of origin. To investigate this, we performed exome sequencing and mutational calling in 5 DLBCL cell lines, followed by mutational calling using total RNA sequencing in the same cell lines and their EVs (Figure 6; supplemental Table 4). We used exome sequencing-based mutational calling as a gold standard and used that dataset to optimize RNA-based mutation calling approach (see “Materials and methods”). Interestingly, RNAs in EVs included a significant number of variants found in the cell of origin as determined from RNA sequencing. Even though EVs contained only a small subset of RNA species compared with whole cells, nearly one-third of all mutations detected in the cell of origin were also identified in EVs, suggesting that mutated RNAs may be specifically packaged in EVs. As an example, OCI-Ly1 cell line carries 451 mutations based on exome analysis, and 79 of those are expected to be present in EVs based on the presence of corresponding RNA species: our analysis detected 71 expected and 4 novel mutations in OCI-Ly1 EVs. The few mutations that are identified only in EVs are due to the low depth of coverage during sequencing: we examined all reads from RNA sequencing and confirmed that all mutations identified in EVs are also present in the cell of origin, but were filtered out during analysis due to low coverage. Low depth of coverage also explains the small number of mutations shared between OCI-Ly7 and corresponding EVs. Identification of shared mutations among lymphoma cells and secreted EVs is the notable finding, which opens new possibilities for disease diagnosis and monitoring in lymphoid malignancies.
Mutational analysis of DLBCL cell lines and EVs. Mutations from DLBCL cell lines are listed in red circles and those from EVs are listed in blue circles with total number of known mutations from exome-seq (green rectangle) demonstrating that several mutations from cell lines are detected in EVs. RNA sequencing indicated that all mutations found in EVs were also present in the cell of origin, but low coverage caused them to be filtered out in the analysis. This technical issue also explains the small number of mutations shared between OCI-Ly7 cells and corresponding EVs, although this could reflect a difference in biology between cell lines. The mutations shared between cell lines and EVs could ultimately provide a way in which disease status could be monitored via liquid biopsy in DLBCL patients.
Mutational analysis of DLBCL cell lines and EVs. Mutations from DLBCL cell lines are listed in red circles and those from EVs are listed in blue circles with total number of known mutations from exome-seq (green rectangle) demonstrating that several mutations from cell lines are detected in EVs. RNA sequencing indicated that all mutations found in EVs were also present in the cell of origin, but low coverage caused them to be filtered out in the analysis. This technical issue also explains the small number of mutations shared between OCI-Ly7 cells and corresponding EVs, although this could reflect a difference in biology between cell lines. The mutations shared between cell lines and EVs could ultimately provide a way in which disease status could be monitored via liquid biopsy in DLBCL patients.
Discussion
Given the complexity of DLBCL pathogenesis and recent reports about biological significance of EVs, we sought to further elucidate their role in DLBCL. We focused on DLBCL because of the poor outcomes of many patients diagnosed with this most common type of lymphoma, and because of the function EVs play in other advanced, high-proliferating cancers.11,12,35 We began by isolating EVs from DLBCL cell lines and showing that they express the exosomal surface marker CD63 and exosomal proteins Alix, TSG101, and CD81, as well as lineage markers. This finding confirms that EVs can be traced from the cell of origin as has been shown before in different lymphoma cell lines.36 Furthermore, we demonstrated that DLBCL cell lines can uptake EVs and their RNAs. This observation highlights the importance of EVs: they promote exchange of aberrant biological programming and thus potentially have a functional impact on recipient cells, including in disease dissemination, as has been demonstrated in melanoma, pancreatic cancer, and chronic lymphocytic leukemia.11,14,37 The possible effects can vary from those on transcriptional program, cell proliferation, and even drug resistance and tumor evolution.
We were particularly interested in the RNA content of DLBCL-derived EVs given its potential to substantially impact the behavior of recipient cells, and because this area has been underexplored. We optimized methods of RNA extraction from EVs to focus on small RNAs that are typically found in EVs. We found that the majority of transcripts found in cell line–derived EVs were shared with the cell lines and those derived from primary DLBCL were shared with the primary sample. Interestingly, RNAs from EVs contain species that reflect the transcriptional program of the cell of origin: we detected that oxidative phosphorylation pathways are overrepresented in EVs derived from ABC DLBCLs. This suggests that EVs can be profiled in order to understand the aberrant transcriptional program associated with disease, and used for targeted treatment. Enrichment of RNAs representing such biological functions as molecular transport and cell-to-cell signaling and interaction points to the possibility that DLBCL EVs may also function to affect tumor microenvironment and prepare niches for tumor spread and homing. All samples contained snRNAs, which play an unknown function in EVs. We recognize that further studies using experimental models and primary samples are needed to develop a comprehensive understanding of the role of these RNAs from EVs. To our knowledge, our study is the first to report RNA sequencing of EVs from DLBCL cell lines and patient sample.
The most intriguing finding from our analysis is that RNAs from EVs can be queried to understand the mutational profile of the tumors that produce them. Given that EVs circulate through the body and can be extracted from peripheral blood, we anticipate that analysis of EVs could provide important information for disease monitoring and guide therapeutic decision making in DLBCL. This approach is advantageous when compared with analysis of circulating tumor cell or cfDNA because the origin of EVs can be delineated based on surface markers, in contrast to cfDNA. Another important aspect in favor of EV mutational profiling is that EVs contain “expressed” RNA species and as such give us clues about aberrant transcriptional programming of the tumor, which can then be targeted. We also showed that EVs seem to preferentially package RNAs that carry mutations and as such may serve yet an unknown function. It is possible that specific mutations in EV-derived RNA could play a key role in disease progression, for example. That feature also allows sampling mutated RNAs that are expressed in the tumor with many likely coding for neoantigens. If this finding can be validated in the larger studies, it may lead to targeted profiling of tumor neoantigens using liquid biopsy. An assay would work as follows: at diagnosis of DLBCL, tumor biopsy and EVs isolated from peripheral blood of the patient would be subjected to mutational analysis (using RNA sequencing or a combination of exome on a biopsy/RNAseq on EV RNA) in order to identify the spectrum of mutations specific to the patient. Once patient-specific mutations are identified, at various points during the treatment and follow-up, peripheral blood would be collected and EVs extracted to assess for the presence of mutations and to monitor disease. We anticipate that this approach may eliminate the need for tissue biopsy and will be superior for monitoring minimal residual disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contributions: R.S. and D.B. designed the research and provided oversight; K.L.R. provided expertise and oversight regarding study design; S.C.R. and A.A.F. designed the experiments and performed the EV isolation and characterization, flow cytometry, and EV exchange assays; A.A.F. also performed confocal imaging and pathway analysis; S.S., A.M., P.M.D., E.C.L., and F.C. assisted with these experiments; A.S. provided expertise on experimental methodology; J.I. extracted RNA from EVs; X.A. sequenced RNA; N.C. and D.B. performed RNAseq analysis; Y.J. analyzed mutations in cell lines; S.L. analyzed mutations in EVs; S.C.R., A.A.F., and R.S. wrote the paper with input from all authors; and all authors read and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Y.J. is Genentech, Inc., South San Francisco, CA.
The current affiliation for J.I. is The New York Genome Center, New York, NY.
Correspondence: Rita Shaknovich, Cancer Genetics Inc, 201 NJ-17, Rutherford, NJ 07070; e-mail: rshaknovich@gmail.com; and Doron Betel, Institute for Computational Biomedicine, Weill Cornell Medicine, 525 East 69th St, Building C, Room 620C, New York, NY 10021; e-mail: dob2014@med.cornell.edu.
REFERENCES
Author notes
S.C.R. and A.A.F. contributed equally to this study.
D.B. and R.S. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal