Abstract
Background
Acute myeloid leukaemia (AML) is a tremendously heterogeneous clonal disorder of haemopoietic progenitor cells and is the most common malignant myeloid disorder in adults. Identified mutations from genomic data cannot provide information about their therapeutic significance without functional data. Hereby, we applied a pooled shRNA library screen to identify the activated signalling pathways essential for the survival of AML cells.
Methods
Mononuclear cells from seven karyotypically normal AML patients were separated from peripheral blood or bone marrow aspirate at diagnosis and transduced with a pooled shRNA library containing 27500 shRNAs targeting 5000 individual genes (Human Module 1, Decipher, Cellecta). The targeted genes were components of known signalling pathways. At 72h post transduction, 30% of the cells were stored for baseline measurements and the rest were co-cultured with HS-5 stromal cells for the selection period. DNA was then extracted and the shRNA barcodes were sequenced on the Illumina NextSeq platform. The frequency of barcode appearance after the selection period was compared to their prevalence at baseline to calculate the shRNA depletion. The shRNAs were filtered to identify those genes which had multiple shRNA meeting a threshold depletion.
Results
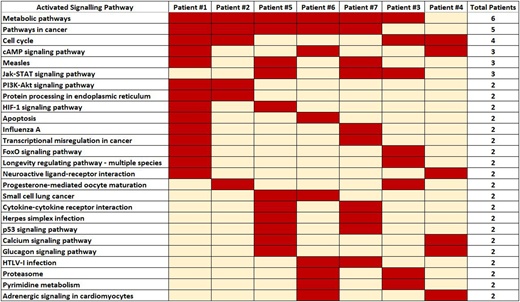
Our data analysis of the 7 AML samples identified various signalling pathways for each of these patients. These data support the notion of heterogeneity in AML. The top 100 depleted genes (with depletion in at least 3 shRNA per gene) in each patient were selected and compared. Our limited initial data showed there to be several activated signalling pathways for each AML sample indicating that inhibition of more than one gene or pathway might be required for efficiently suppressing these leukaemia cells.
Common targets: NOX1 was the most commonly identified therapeutic target among the screened patients being significantly depleted in AML cells from 5/7 patients. This is an important finding as there are available NOX1 inhibitors for treatment of colon cancers and can be investigated as a therapeutic option for acute myeloid leukaemia. The other most common targets were CDK5R1, DISC1, FSCN3, and PSMB7 which were found to be significantly depleted among 3 of the 7 screened patients. The merged data also showed 58 essential genes for AML cell survival were common in at least 2/7 patients. Using Enrichr the activated signalling pathways based on the top selected genes were identified. Various signalling pathways were observed for each patient showcasing the heterogeneity among AML patients (Figure 1). However, some signalling pathways were indeed common among multiple patients - with different genes being responsible for the activation of those pathways among the patients. The most common pathway was the metabolic pathway which was observed among the top 20 essential pathways in 6/7 patients. The JAK-STAT5 signalling pathway, purine metabolism and cAMP signalling pathway were also among the top 20 essential pathways in 3/7 patients while the following pathways: FoxO, PI3K-AKT, HIF-1, P53, Glucagon, and proteasome were observed in 2/7 patients. Identification of several essential survival pathways provides the opportunity to develop personalised therapy through combined targeting of more than one pathway.
Conclusion
The signalling pathways analysis using candidate genes from a pooled shRNA library screen showed patient-specific signalling pathways and also common pathways among these screened patients. Absence of a common gene among the screened patients further highlights the significance of personalised therapy in AML and the necessity of developing diagnostic tools to identify potential targets at diagnosis. Identification of crucial genes such as NOX1 (a gene known to have a role in the survival of leukemic stem cells) and other genes with known significance in the pathogenesis of AML supports the application of this method for identifying therapeutic targets at diagnosis or relapse.
Knapper:Jazz: Other: Meeting and travel support; Daiichi Sankyo: Other: Meeting and travel support; Chroma Therapeutics: Research Funding; Celgene: Other: Meeting and travel support; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees. Apperley:Pfizer: Honoraria, Speakers Bureau; Incyte: Honoraria, Speakers Bureau; Novartis: Honoraria, Research Funding, Speakers Bureau; BMS: Honoraria, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal