Abstract
Background: Multi-parameter flow cytometry (MFC) data is a diagnostic keystone for hematologic malignancies. Data analysis and interpretation requires resources and expertise, full automation is desired. Combination of automated gating strategies and machine learning on MFC data proved effective in detecting AML cell populations vs normal (Aghaeepour et al., 2013). Aim of our approach was to create tools capable of comprehensively analyzing and classifying MFC data. We developed a proof of principle framework to process MFC data generated from samples with suspected B-cell lymphomas.
Methods: MFC data was obtained from routine diagnostics of 16,384 patients as follows: normal controls (n=8,493), chronic lymphocytic leukemia (CLL, n=3,412), CLL with increased prolymphocytes (CLL/PL, n=603), follicular lymphoma (FL, n=219), hairy cell leukemia (HCL, n=193), lymphoplasmacytic lymphoma (LPL, n=629), mantle cell lymphoma (MCL, n=269), marginal zone lymphoma (MZL, n=979), monoclonal B-cell lymphocytosis (MBL, n=1,480). Two 9-color combinations of monoclonal antibodies were applied to analyze surface expression of the following antigens besides scatter signals: FMC7, CD10, IgM, CD79b, CD20, CD23, CD19, CD5, CD45, Kappa, Lambda, CD38, CD25, CD11c, CD103, CD22. For dimensionality reduction we used an unsupervised clustering approach (Kohonen et al., 1990). We first generated a consensus self-organizing map (CS) of 100 nodes including all MFC events of multiple preselected cases (5 samples/cohort). For a test case the fluorescent profile of a cell was assigned to its nearest node in the CS (upsampling). We used the resulting distribution for classification by a sequential neural network (NN). Computationally intensive processing steps were run using Amazon Web Services. Mean accuracies were calculated counting individual cases across the entire dataset. All accuracies are described for 10 CS generations and 10-fold cross validation in the classification process. Approaches of data processing were sequentially developed aiming at maximization of classification accuracy.
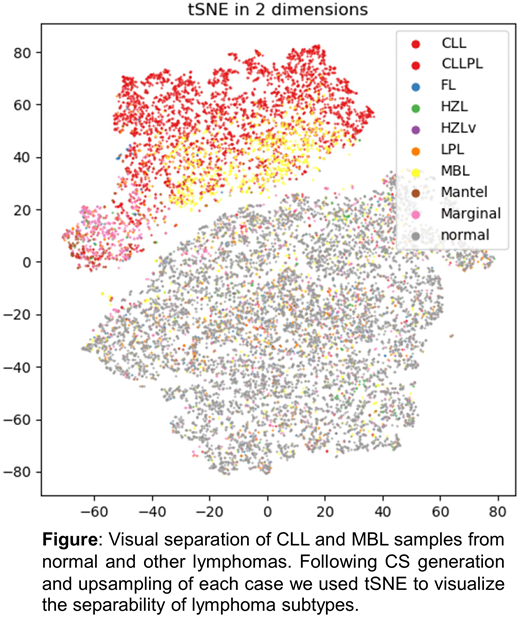
Results: First, we assessed the performance in a binary classification, i.e. the merged group of CLL and MBL (n=4,877) vs. normal (n=8,493) achieving 97% accuracy. Secondly, we applied our approach to the classification of all 9 classes with an accuracy of 74%. Visual inspection of tSNE (van der Maaten et al., 2008) transformed upsampling information revealed good separability between CLL and normal cases but substantial overlap between other lymphoma subtypes (Figure). The following approaches were applied to improve classification:
1) Automated pregating of lymphocytes (CD45/SSC) by DBSCAN analysis (Ester et al., 1996) improved classification. Applying this to both CS generation and upsampling the accuracy of the 9-class problem increased to 78% (p<0.001).
2) Using a CS trained only on cases with unequivocal phenotypes without pregating was significantly better (p=0.01), but pregating overrode this effect (p=0.5).
3) According to the partial overlap of immunophenotypes hierarchical clustering of misclassifications showed overlap mainly between MCL and CLL/PL as well as between LPL and MZL. We therefore changed our approach merging the groups CLL/MBL, MCL/CLL/PL and LPL/MZL while keeping FL, HCL and normal controls separate. Automated classification of these 6 classes resulted in 86% accuracy.
4) Further regrouping into 3 classes, according to the commonly applied routine approach of distinguishing normal cases from CD5+ (CLL, MBL, MCL, CLL/PL) and CD5- B-cell lymphoma (FL, LPL, MZL, HCL) resulted in a classification accuracy of 89%.
5) The misclassified cases in the 3-class experiment had an average infiltration of 10% versus 41% for correctly classified ones. Limiting the automated classification to cases with ≥5% infiltration increased the classification accuracy to 95%.
Conclusion: We created a framework for automated classification and analysis of routine diagnostic MFC data. Definition of entities and preselection of cell populations are essential for optimizing classification accuracy. Our approach is the first application of computational MFC and AI on a large scale dataset and proves the feasibility in a routine diagnostic work flow. To further improve the accuracy of classification a future algorithm will integrate distance metrics following SOM generation into the NN classifier.
Höllein:MLL Munich Leukemia Laboratory: Employment. Schabath:MLL Munich Leukemia Laboratory: Employment. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal