Abstract
Background
Extensive sequencing data highlighted the recurrence of mutations resulting in constitutive MAP kinase signaling in chronic lymphocytic leukemia (CLL), and proposed these mutations as late drivers of CLL progression (X.S. Puente, Nature, 2015; D.A. Landau, Nature, 2015). Although some preliminary studies suggested KRAS, NRAS and BRAF mutations as possibly associated with trisomy 12 (TRI12) and/or unmutated (UM) IGHV genes (C.D. Herling, Blood, 2016; K. Takahashi, Blood, 2018), dedicated analysis of these aspects is still missing.
Aim
To correlate the occurrence of KRAS, NRAS and BRAF mutations with specific clinico-biological features in CLL.
Methods
The study included 534 primary CLL purposely enriched with 300 TRI12 cases: 190 cases with TRI12 as sole chromosomal aberration (TRI12only) and 110 cases harboring TRI12 plus other chromosomal aberrations (TRI12plus). The cohort was also enriched in cases with NOTCH1 aberrations (n=214) to evaluate their possible role in RAS-RAF mutations incidence.
Amplicon-based targeted next-generation sequencing assay was performed using Miseq Illumina on DNA from purified CLL samples. Analyzed amplicons mapped in the hotspot regions of KRAS, NRAS (exons 2, 3 and 4) and BRAF (exons 11 and 15) genes. Variant allele frequency (VAF) was obtained by Miseq report. CLL cases were characterized for IGHV mutational status, main cytogenetic abnormalities, NOTCH1 and TP53 aberrations. Time to first treatment (TTFT) data were correlated with molecular findings.
Results
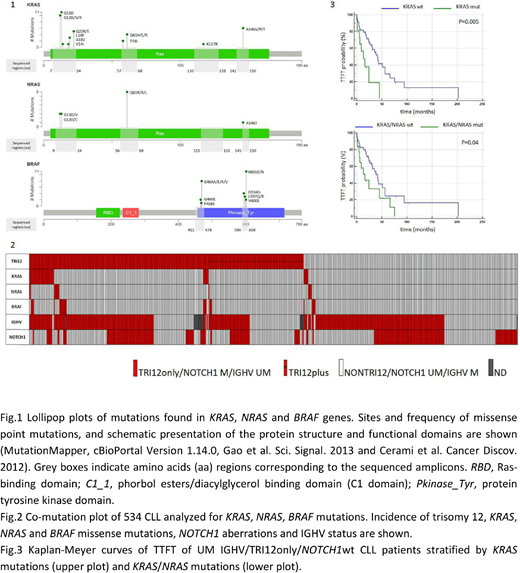
We found 90 missense point mutations (Fig.1) in 64 CLL cases, with prevalence of KRAS (44 mutations in 38 patients), followed by BRAF (31 mutations in 24 patients) and NRAS (15 mutations in 13 patients). Nearly all mutations have been previously associated with gain-of-function phenotype and increased RAS/ERK downstream signaling. The co-occurrence of 2 mutated genes (mainly KRAS and BRAF) were observed in 11 patients. Mutations were mainly subclonal (median VAF 6.15%, range 1.3%-61.6%) with only one third of mutations (27/90) above 15% VAF.
A high association between the presence of KRAS/NRAS/BRAF mutations and UM IGHV and the presence of TRI12 was observed. Overall, 87.3% of KRAS/NRAS/BRAF mutated cases had a UM IGHV (p<0.0001) and 79.7% were TRI12 CLL (p<0.0001) (Fig.2). Moreover, in the context of the UM IGHV CLL group (332 cases), TRI12 CLL had a 4-fold higher KRAS/NRAS/BRAF mutation frequency (46/182, 25.3%) than non-TRI12 (9/150, 6%; p<0.0001). Indeed, we found 29/182 (15.9%) KRAS, 9/182 (4.9%) NRAS and 17/182 (9.3%) BRAF mutated cases in the TRI12 group vs. 4/150 (2.7%) KRAS, 3/150 (2%) NRAS and 3/150 (2%) BRAF mutated cases in the non-TRI12 group. The KRAS/NRAS/BRAF mutation frequency was even higher when considering the TRI12only group in the context of UM IGHV (38/133, 28.6%). Conversely, CLL cases with del(13q) as sole chromosomal aberration (n=94) showed the lowest frequency of KRAS/NRAS/BRAF mutations (2/94, 2.1% in the whole cohort and 2/53, 3.8% in the context of UM IGHV cases).
Furthermore, a higher prevalence of KRAS/NRAS/BRAF mutations was found in NOTCH1 wild type (wt) (22.1%) when compared to NOTCH1 mutated (11.2%) cases, in the context of the UM IGHV group (p=0.008), pointing to a mutual exclusivity of these mutations in the pathogenesis of disease.
No other significant associations with clinical variables as RAI stage at diagnosis, presence of TP53 mutations or gender were observed.
We finally evaluated if KRAS/NRAS/BRAF mutations had an impact on TTFT in the context of UM IGHV/TRI12only/NOTCH1wt, that showed the highest incidence of mutations (26/70, 37%). Both KRAS mutations alone (p=0.005) and KRAS/NRAS mutations (p=0.04) were associated with shorter TTFT (Fig.3).
Conclusions
Taken together our data show that KRAS, NRAS and BRAF mutations are almost exclusively found in UM IGHV/TRI12/NOTCH1wt CLL and, in the context of this CLL group the presence of KRAS and NRAS mutations is associated with unfavorable prognosis.
The high incidence of KRAS, NRAS and BRAF activating mutations in TRI12 CLL together with the observation of higher level of ERK phosphorylation (S. Decker, Blood, 2012) and higher sensitivity to MEK/ERK inhibitors (S. Dietrich, J Clin Invest, 2018) described in the TRI12, reinforce the evidence of an essential role for MEK/ERK signaling in TRI12 CLL, pointing to this pathway as the most appropriate therapeutic target.
Zaja:Novartis: Honoraria, Research Funding; Janssen: Honoraria; Abbvie: Honoraria; Celgene: Honoraria, Research Funding; Amgen: Honoraria; Takeda: Honoraria; Sandoz: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal