Abstract
Although a pathogenic role for the Epstein-Barr virus (EBV) is largely undisputed for tumors that are consistently EBV genome positive (eg, nasopharyngeal carcinoma, endemic Burkitt lymphoma), this is not the case for classical Hodgkin lymphoma (cHL), a tumor with only a variable EBV association. In light of recent developments in immunotherapeutics and small molecules targeting EBV, we believe it is now timely to reevaluate the role of EBV in cHL pathogenesis.
Classical Hodgkin lymphoma
Classical Hodgkin lymphoma (cHL) is characterized by rare malignant Hodgkin/Reed Sternberg (HRS) cells in an inflammatory tumor microenvironment (TME). HRS cells are B cells with evidence of somatic hypermutation indicating that they are germinal center (GC) experienced.1 However, phenotypically, HRS cells do not resemble B cells and lack a functional B cell receptor (BCR); in ∼25% of cases, this is caused by mutations that destroy the coding capacity of the immunoglobulin genes.2
Risk for cHL in people infected with EBV
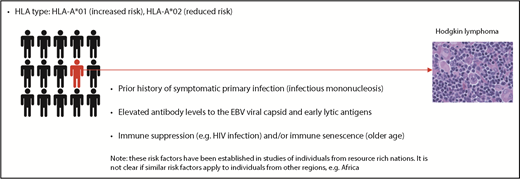
The detection of raised antibody levels to EBV antigens in cHL patients provided the first evidence that EBV might be involved in cHL.3 These raised levels precede cHL development,4 an effect specific to virally associated cases (Figure 1).5 People with a prior history of infectious mononucleosis (IM) also have an increased cHL risk6,7 that is specific to EBV+ cases.8,9 EBV+ cHL is temporally related to primary infection, developing a median of 2.9 years after IM.8 In children, EBV+ cHL, but not EBV− cHL, is associated with a seasonal peak in the onset of symptoms, consistent with an increased risk for the tumor following primary infection.10
An increased and decreased risk of EBV+ cHL is also associated with HLA-A*01 and HLA-A*02 alleles, respectively.11-15 Although the same genotypic markers that confer a risk for EBV+ cHL are associated with IM risk, HLA-A typing has revealed that a prior history of IM is independently associated with EBV+ cHL, even after adjusting for the effects of HLA-A alleles.
The risk factors associated with cHL have been identified almost exclusively in individuals from resource-rich nations. However, we do not know whether these risk factors are the same for cHL occurring in resource-poor countries (eg, in sub-Saharan Africa where IM is rare [>80% of children are infected at younger than 5 years of age]16 but 70% to 90% of cHL is EBV+).17,18
Detection of EBV in cHL tissues
Infection of the tumor cells is considered a sine qua non for establishing a causal association between virus and tumor. In cHL, these conditions were fulfilled shortly after the initial serological analyses had implicated EBV. Thus, in 1985, the EBNA protein (Epstein-Barr nuclear antigen, now known to be EBNA1) was detected in HRS cells.19 EBV DNA was detected in 20% to 25% of Hodgkin lymphoma biopsies by Southern blotting,20 and in situ hybridization for EBV DNA confirmed the EBV genome in HRS cells.21,22 Later, the abundant untranslated Epstein-Barr encoded RNAs (EBER1 and EBER2) would provide a target for the routine detection of EBV in tumors, including cHL.23 Finally, HRS cells were shown to express the EBV-encoded latent membrane protein-1 (LMP1), LMP2A, and EBNA1 proteins.24-27
Viral genomes are monoclonal in cHL. Upon cell entry, the linear EBV DNA circularizes by recombination of the terminal repeats (TRs), producing fused termini of a unique length for each circularization event. Detection of multiple TR lengths within a sample specifies a polyclonal infection, whereas TRs with an identical number of repeats indicate expansion of a single infected progenitor cell. Thus, the detection of a single TR length in cHL strongly argues for an etiological role for EBV.21 Moreover, the number of EBV genomes present per cell is different between patients but is relatively constant in different samples from the same patient.28 EBV also persists in HRS cells throughout the course of disease and is present in multiple sites of disease28 (Figure 2). These are important observations; EBV episomes can be lost by asymmetrical segregation during cell divisions with retention of EBV genomes in cHL suggesting that the virus provides an ongoing advantage to the tumor cells.
The frequent detection of EBV in cHL and, in these cases, the presence of the virus in every tumor cell is strong evidence of an etiological association. EBV is present in a third of cHL cases in the west; the EBV association is even higher in some parts of the world. Were the virus simply a passenger, then the frequency of EBV+ cHL would approximate the number of EBV-infected B cells in an individual (ie, ∼1 in 106 of the total B cell pool). If we repeat this calculation, this time assuming that EBV+ cHL arises only from GC B cells, the expected frequency of EBV+ cHL would be equivalent to the frequency of EBV-infected B cells in the GC (ie, 4.6 per 104, using the highest reported frequency of EBV infection in GC B cells).29
Evidence of a causal role for EBV from epidemiological studies
The geographical patterns of cancer incidence can indicate an infectious etiology. Resource-rich nations generally have a lower burden of virally related cancers than resource-limited countries.30,31 Thus, EBV rates in cHL from North America and Europe are lower than in resource-limited countries.32,33 EBV+ rates for cHL are also higher in Asians and Hispanics compared with whites or blacks,30 as well as in South Asian children compared with non-South Asian children in the UK.34 In resource-rich countries, the proportion of cases with EBV is highest in older people and children, with lower rates in young adults.35,36 The global patterns of EBV+ cHL differ somewhat from those observed for EBV+ Burkitt lymphoma which is characterized by very high rates in so-called “endemic” areas but much lower rates across the rest of world.
Virus-associated cancers are increased in immunosuppressed persons (exemplified by posttransplant lymphoproliferative disease; PTLD) and in older people, an effect attributed to an age-related decline in virus-specific immunity.35 However, EBV+ cHL is the least common major form of PTLD. Moreover, although the incidence of EBV+ cHL is increased in HIV infection,37,38 this is only observed in patients with intermediate levels of immune impairment.37 Moreover, the incidence of cHL in HIV+ patients has not fallen in the era of highly active antiretroviral therapy; indeed, cHL risk may be increased following immune reconstitution on highly active antiretroviral therapy.39-41 This apparent paradox can be explained by the dependency of HRS cells on CD4+ T cells.
EBV latent genes contribute to cHL pathogenesis
Three EBV-encoded proteins and viral microRNA (miRNA) expressed in virus-infected HRS cells are implicated in cHL pathogenesis.
EBNA1 is essential to maintain EBV infection, acting as a viral replication factor and tether between the viral genome and the host cell chromosomes ensuring (for the most part) faithful segregation of viral episomes during cell division.42 EBNA1 is also a transcription factor that regulates viral and cellular gene expression.43-45 EBNA1 can promote the growth and survival of HRS cells, an effect attributable to the downregulation of the protein tyrosine phosphatase receptor type K.46,47
LMP1 is a constitutively active CD40 receptor homolog48 that induces many of the gene expression changes characteristic of HRS cells.49-51 LMP1 induces multiple cell signaling pathways that are aberrantly activated in HRS cells, including NF-κB and JAK/STAT.52-55 Alternatively, these pathways can be activated by cellular mutations in cHL.56 Accordingly, EBV+ cHL has significantly fewer cellular mutations,57 including mutations in negative regulators of the NF-κB pathway (eg, TNFAIP3, NFKBIA).58-60 EBV+ tumors also have fewer chromosomal breakpoints and aneuploid autosomes.61 Thus, in EBV+ cHL, viral genes are thought to be sufficient to drive aberrant activation of signaling pathways in the absence of cellular mutations, indicating that EBV and cellular mutations provide mutually exclusive means to the same pathogenic end point.
A variant of LMP1, carrying a 30–base pair deletion in the C terminus, reported to have enhanced transforming properties62,63 has been found more commonly in HIV+ cHL compared with non-HIV–associated cHL,64 as well as in pediatric cHL compared with normal controls.65 A study of HIV patients also identified LMP1 polymorphisms associated with increased NF-κB activation, but these variants were not found to be significantly enriched among EBV+ cHL.66
LMP2A is a BCR mimic that allows B-cell development in the absence of normal BCR signaling.67,68 EBV can immortalize BCR− GC B cells, an effect that is dependent upon LMP2A.69-72 Consistent with this, crippling mutations in immunoglobulin genes are found almost exclusively in EBV+ cHL.73 LMP2A also suppresses expression of B-cell transcription factors, including EBF1 and E2A, in turn recapitulating many of the gene expression changes seen in HRS cells.74-77
The EBV latent genes probably also contribute to the characteristic TME of cHL. Thus, LMP1 can recruit cells of the TME by inducing the expression of numerous chemokines and cytokines in infected HRS cells.78-80 Similarly, EBNA1 has been shown to increase expression of the T-cell chemokine CCL20 and, in turn, the recruitment of regulatory T cells.81
Loss of HLA expression is common in HRS cells, yet, counterintuitively, HLA class I loss is more frequent in EBV− cHL,82-84 which, in nodular sclerosis cases, could be due to mutations in the β-2 microglobulin gene.85 In contrast, EBV+ cHL expresses normal or even elevated levels of HLA and contains more activated cytotoxic T lymphocytes than EBV− cHL.82-84,86 Other mechanisms could explain immune evasion in EBV+ cHL. For example, LMP1 has been shown to induce PD-L1 expression to suppress cytotoxic T cell responses.87
EBV-encoded BART miRNAs are also expressed in EBV+ cHL88 and could contribute to lymphomagenesis by regulating viral and cellular gene expression; BART miRNAs have effects on innate immunity (eg, miR-BART6-3p targets DDX58 encoding retinoic acid–inducible protein 1, a sensor of viral messenger RNA),89 adaptive immunity (eg, miR-BART1 and miR-BART2, which reduce antigen presentation, and miR-BART17, which targets TAP2),90,91 apoptosis (eg, multiple BART miRNAs targeting caspase 3),92,93 and proliferation (eg, miR-BART6-3p targets PTEN).94
Future prospects to incorporate EBV testing and targeted therapies in the management of cHL
The detection of EBV within cHL, although sometimes useful in confirming the diagnosis, does not influence patient management. Combination chemotherapy and radiotherapy is the mainstay of treatment of cHL patients, with 5-year survival rates in the range of 80% to 90%. However, the prognosis for patients with refractory disease or for those who relapse after conventional therapy is dismal. The current treatments also cause significant long-term toxicities that include secondary malignancies, cardiopulmonary toxicity, hypothyroidism, and infertility. Outcomes are particularly poor for older patients (5-year survival 30-50% in patients older than 70 years).95 Furthermore, an EBV+ status in elderly patients is associated with an even worse prognosis.96-100
It is in this context that immune checkpoint inhibitors targeting the PD-1 pathway are providing impressive clinical results, even in heavily pretreated patients.87,101-104 Further improvements in these clinical responses could be achieved by combining immune checkpoint inhibitors with EBV-specific therapies. Ex vivo–manufactured donor T cells and patient-derived EBV-specific T cells are already available and have proven effective, particularly in patients with PTLD.105,106 More recently, vaccine approaches have been developed based on recombinant virus proteins or virus peptides.107,108 Furthermore, recent developments in the identification and design of EBNA1 inhibitors have opened up new possibilities to therapeutically target EBV-associated malignancies, including cHL.109-112
Conclusions
Although some cHL patients harbor EBV in tumor cells, the contribution of the virus to this tumor has been the subject of debate. Differences in the epidemiology, genetics, biology, and clinical behavior of EBV+ vs EBV− cHL strongly indicate a pathogenic role for the virus. A detailed understanding of the relationship between EBV and cHL provides insights into cHL pathogenesis and highlights opportunities for novel therapeutic interventions.
Acknowledgment
This work was supported by Bloodwise.
Authorship
Contribution: P.G.M. and L.S.Y. conceived the idea and wrote the manuscript.
Conflict-of-interest disclosure: L.S.Y. is a member of the Scientific Advisory Board of Viracta Therapeutics, Inc. P.G.M. declares no competing financial interests.
Correspondence: Paul G. Murray, Institute of Immunology and Immunotherapy, University of Birmingham, Vincent Dr, Birmingham B15 2TT, United Kingdom; and Health Research Institute, University of Limerick, Limerick V94 T9PX, Ireland; e-mail: p.g.murray@bham.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal