B-other ALL represents a working definition for patients with B cell precursor (BCP) ALL without a known primary chromosomal abnormality. In this study we use whole genome sequencing (WGS) to characterize adult B-other cases (age ≥ 25yrs) from the UKALL14 trial (NCT01085617).
Of 652 patients aged 25-65yrs enrolled onto UKALL14, 333 (51%) had B-other ALL. Sufficient material was available to screen 156/333 B-other cases for recurrent Ph-like fusion events (CLRF2, JAK2, ABL1, ABL2PDGFRB) using FISH and MLPA (kit P335). This identified 28 (18%) Ph-like fusion events (21 CRLF2, 5 ABL-class fusions and 2 JAK). Of the remaining 128 B-other cases 57 had available samples for tumor normal paired WGS (read depth 60x and 30x respectively). Bioinformatic analysis was performed to determine small somatic mutations (SSMs); single nucleotide variants (SNVs) and insertion/deletions (INDELs) as well as copy number aberrations (CNA) and structural variants (SV). We also undertook de novo motif analysis to identify RAG mediated deletions.
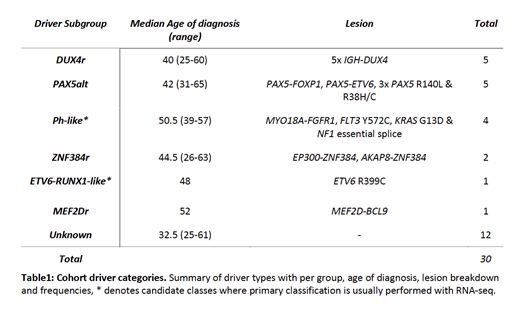
We present data for 30/57 cases (median age of diagnosis 44, range 25-65), the remainder are undergoing sequencing. Within this cohort we identified 784 SVs, 49,244 SNVs and 2,881 INDELs, with each case having a median representation of 23 SVs, 1,674 SNVs and 86 INDELs. The Median SSM burden was 0.55 per megabase (range 0.31-0.81), which is in the upper third of previous ALL estimates (median 0.26 range 0.03-2.9) but low compared to most other cancer types (Alexandrov et al. 2013 Nature).
Fusion gene analysis identified 11/30 (37%) cases with recurrent rearrangements (2 ZNF384r, 2 PAX5r, 5 DUX4r and FGFR1r); this identified a novel 5' partner for ZNF384r (AKAP8) and MYO18A-FGFR1 a fusion previously seen in a single case of 8p11 myeloproliferative syndrome. A further 7 clonal coding drivers were identified, 3 PAX5alt, 3 Ph-like candidates and a single ETV6-RUNX1-like candidate (ETV6 R399C, a dominant negative mutation).
We classified our cohort into 6 driver subgroups as shown in table 1 using Gu et al. as a framework (Gu et al. 2019 Nat Gen). We found no evidence of driver subgroups clustering with age, although the Ph-like candidates were all identified in patients 39yrs or older. There was evidence that known driver subgroups have differing mutation burdens but these were not significant in this preliminary cohort; the single MEF2Dr case had a high SV burden compared to all other known subgroups (137 vs. cohort median 22); Favorable outcome subgroups (ZNF384r and DUX4r n=7) had a lower SNV coding mutation burden (median 14 vs. 20; U = 60, p = 0.057; Mann-Whitney U test); PAX5alt (n=5) cases had a higher INDEL burden (median 174 vs. 82; U = 16.5, p =0.057). As expected we found recurrent CNA in CDKN2A/B (63%; 19/30), PAX5 (33%; 10/30), IKZF1 (27%; 8/30), ETV6 (11%; 3/30) and BTG1 (7%; 2/30) across the cohort. IKZF1 deletions were enriched in the Ph-like candidate (n=4) subgroup compared to all other known groups (75% vs. 12%; p = 0.028; Fisher's Exact).
RAG mediated deletions have been established as an oncogenic driver mechanism in childhood ALL, so we sought to ascertain its role in adults. We identified 54% (128/236) of the breakpoint resolved deletions had a RAG heptamer site. There was a significant difference (U = 167.5, p = 0.021) in RAG deletion burden between patients over 40yrs and under 40yrs at diagnosis (median 1.5 vs 3).
Within the unknown driver subgroup; three cases carried ZEB2 H1038R mutations, two with concurrent IGH-CEBPA/B fusions, identifying them as likely belonging to the G12 cluster identified by Li et al. (Li et al. 2018 PNAS); one case had a high translocation burden (37 cohort median 2) and a single case had a high RAG deletion burden (45, cohort median 2); of the remaining 7 cases, four had either a NRAS or FLT3 subclonal hotspot mutation.
Here we present the WGS analyses of 30 cases classified as B-other on the basis of no cytogenetic findings by standard clinical assays. The landscape of adult B-other ALL is highly heterogeneous but with WGS we were able to find at least one disease defining event in 60% of our cohort. These events often encompass novel fusion partners of established genomic subtypes or a cytogenetically cryptic lesion such as IGH-DUX4. Taken together our findings demonstrate the clinical utility of WGS as a diagnostic assay to inform and improve the management of adult B-other ALL patients in the future.
Fielding:Pfizer: Consultancy; Incyte: Consultancy; Amgen: Consultancy; Novartis: Consultancy. Papaemmanuil:Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal