BACKGROUND: Older patients with acute myeloid leukaemia (AML) who are unsuitable for standard induction therapy have limited treatment options. While DNMT3A, TET2, IDH1/2 and TP53 mutations have been previously associated to better response to hypomethylating agents, there are no molecular biomarkers for low-dose cytarabine (LDAC)-based regimens.
AIMS: To predict outcome in AML older patients at diagnosis based on mutation status in the context of FLUGAZA trial. FLUGAZA trial was focus on >65 years AML de novo patients comparing azacytidine vs. fludarabine and LDAC (FLUGA Scheme).
METHODS: We analyzed bone marrow (BM) samples at diagnosis from 209 out of 285 AML patients treated according Flugaza trial (NCT02319135), azacytidine-arm (n=97) and FLUGA-arm (n=112). In this trial, patients were randomized to receive 3 induction cycles with fludarabine and cytarabine (FLUGA) followed by 6 consolidation cycles with reduced intensity FLUGA, vs 3 induction cycles with 5-azacytidine (AZA) followed by 6 consolidation cycles with AZA. Median age at diagnosis was 75 years (65-90). Both treatment groups were balanced for age, leucocytes count, baseline BM blasts, karyotype risk (ELN), and FLT3-internal tandem duplication and NPM1 gene mutations. Mutational profile analysis was carried out by NGS targeted gene sequencing (Ion Torrent S5XL System-Thermo Fisher Scientific) using a 43 genes custom panel implicated in leukemia prognosis.
RESULTS:
We detected 893 variants, 247 Indels and 646 SNVs. 206 (23.1%) of them were included as pathogenic or like-pathogenic by clinvar database. Ninety-five percent of patients (n=203) had at least one detectable mutation, and the median number of mutations was 4 (range = 0-8 mutations). The most common gene mutations were TET2 (N=55), FLT3 (n=52), SRSF2 (n=49), TP53 (n=45), DNMT3A (n=45), ASXL1 (n=45), RUNX1 (n=43), IDH2 (n=36), IDH1 (n=34), NPM1, (n=33) and NRAS (n=23). This mutational landscape is different to previous published in younger patients (Grimwade, Blood 2016), with higher number of patients with mutations in TP53 (21.5 vs 8%), SRSF2 (23.9 vs 2%), IDH1 (16.3 vs 7%) and IDH2 (17.2 vs 9%) and lower number of patients with mutation in NMP1 (15.8 vs 33%).
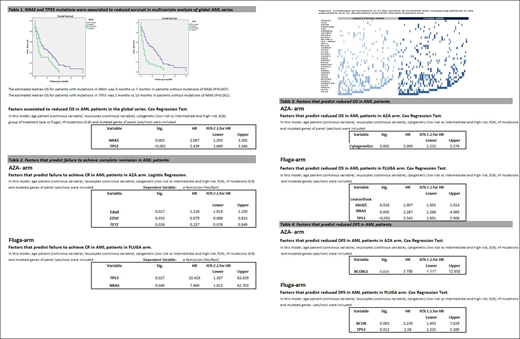
The median OS of global series was 6 months (range 0-40). Multivariate Cox regression in the global series showed that NRAS and TP53 mutations predict reduced OS (Table 1). Distribution of mutations between both arms was not homogeneous (Figure 1) and NRAS (p=0.012) was more frequent among patients randomized to the FLUGA-arm. However, TP53 mutation frequency distribution was homogeneous: 23.7% in AZA-arm and 19.6% in FLUGA-arm (p=NS).
In the AZA-arm, patient´s age was the only variable associated with not achieving composite complete remission (CR plus CR with incomplete recovery) and TET2 and EZH2 mutations were predictors to achieve composite CR. In the FLUGA-arm, TP53 and NRAS mutations were associated with not reaching composite CR (table 2).
In the AZA-arm, cytogenetic was the only variable associated with risk of early death. In the FLUGA-arm, leucocyte count, TP53 and NRAS mutations were associated with risk of early death (table 3).
In the AZA-arm, BCORL1 mutations (4.1%) were the only variable associated with high risk of relapse. In the FLUGA-arm, BCOR (7.1%) and TP53 (19.6%) mutations were associated with high risk of relapse (table 4).
CONCLUSION
The mutational profile of AML in elderly patients is different from the previously published in young patients. We have confirmed that a molecular pattern can identify patients with poor prognosis in elderly AML patients. NRAS and TP53 mutations confer a poor prognosis in LDAC (FLUGA-arm) patients, but this effect disappeared in the AZA-arm. BCOR and BCORL1 mutations were associated to a reduced DFS.
These results confirm that azacytidine could be more efficacious than LDAC treatment for older patients with AML and mutations in TP53, NRAS, TET2 and EZH2. The percentage of patients who presented mutations in these genes amounted to 77% in this AML series.
The study is registered at www.ClinicalTrials.gov as NCT02319135.
This study was supported by the Subdirección General de Investigación Sanitaria (ISCIII, Spain) grants PI13/02387 and PI16/01530.
Salamero:Celgene: Honoraria; Novartis: Honoraria; Pfizer: Honoraria; Daichii Sankyo: Honoraria. Paiva:Amgen, Bristol-Myers Squibb, Celgene, Janssen, Merck, Novartis, Roche, and Sanofi; unrestricted grants from Celgene, EngMab, Sanofi, and Takeda; and consultancy for Celgene, Janssen, and Sanofi: Consultancy, Honoraria, Research Funding, Speakers Bureau. Fernandez:Pfizer: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Celgene: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Teva: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Abbvie: Membership on an entity's Board of Directors or advisory committees; Incyte: Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; Novartis: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Karyopharm: Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Daiichi Sankyo: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal