Background. TP53 mutations (TP53mut) along with 17p13 deletion (del17p) are strong predictors of poor survival and refractoriness to chemo-immunotherapies (CIT) in chronic lymphocytic leukemia (CLL), and their analyses should be always performed before treatment. Studies based upon ultra-deep-NGS have shown that TP53mut can be present at very low level in CLL cell populations, although their detrimental clinical impact in this setting is still matter of debate (Rossi, Blood 2014), and ERIC recomendations discourage to report TP53 mutations if subclonal Malcikova, Leukemia 2018).
Aim. To investigated the presence and clinical relevance of clonal/subclonal TP53 aberrations in a large CLL cohort.
Methods. The study includes 1,058 out of 1,613 CLL patients (509 treated with standard CIT) diagnosed between 1991 and 2018, and consecutively referred to a single institution for del17p analyses by FISH (167-kb 17p13 orange probe, MetaSystems), and TP53mut by ultra-deep NGS (MiSeq Illumina; median coverage >2,000X with an amplicon-based strategy covering exons 2-11 using 40ng DNA/test) in CD19-purified (>85% pure) CLL samples, collected before treatment (as per ERIC recommendations). For TP53mut analyses, FASTQ files were aligned to the Hg19 reference with Burrows-Wheeler Aligner-MEM algorithm, and allele variants called by FreeBayes (Garrison & Marth, arXiv 2102) with non-stringent parameters. To calculate random/systematic errors we generated a specific database with all the variant allele frequencies (VAF) observed in a subset of TP53 wild type (wt) subjects (n=362). TP53mut were accepted if: i) validated by Fisher exact test after Bonferroni correction (p<0.01); ii) with a VAF at least 2.75 standard deviations from the mean of the transformed distributions. The minimal allelic fraction for TP53mut calling was 0.4%. TP53mut cases with less than 2% VAF were tested twice. Outcome was overall survival (OS) from the date of exam.
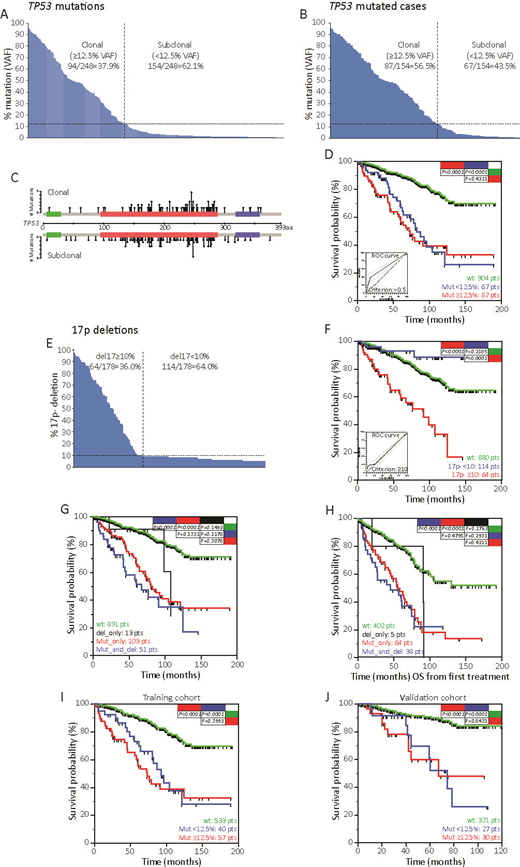
Results. A total of 248 TP53mut (Fig.A) were found in 154 patients (13.5%, Fig.B) with a median mutations/patient of 1.65 (range 1-11). According to the 12.5% VAF cutoff for TP53mut (Nadeu, Blood 2016), 87 cases were clonal (at least one clone > 12.5%) and 67 subclonal (all clones <12.5%; Fig.AB). Clonal and subclonal TP53mut have similar molecular characteristics (Fig.C), supporting the idea of comparable pathogenic effects. In fact, cases bearing clonal and subclonal TP53mut experienced comparably OS, shorter than TP53-wt cases (Fig.D). Accordingly, ROC analysis identified a cutoff of VAF >0.4% for the clinical impact of TP53mut (Fig.D), and the c-index of combined clonal/subclonal TP53mut (0.645) was significantly higher than the c-index of clonal TP53mut alone (0.602; P<0.001). Del17p was found in 178 patients (16.8%; Fig.E). In keeping with a ROC analysis (Fig.F, inset), the 64 cases with del17p in >10% of nuclei had significantly shorter OS than cases with del17p in <10% or without del17p (Fig.F). By combining del17p and TP53mut according to ROC cutoffs, 891 cases (84.2%) presented no TP53 aberrations, 13 cases (1.2%) were del17p only, 103 cases (9.7%) were TP53mut only, and 51 cases (4.8%) were del17p/TP53mut. Compared to TP53-wt cases, similar shorter OS were observed for TP53mut and del17p/TP53mut cases (Fig.G). Same results were obtained in the context of treated patients when OS was computed from the date of treatment (Fig.H). The "yes/no effect" of TP53 mutations on CLL outcome was verified dividing our cohort in a training cohort of 630 CLL cases, and in a validation cohort of 428 CLL patients. These two cohorts presented similar OS, and the same proportion of TP53 mutated cases and 17p deletion. ROC analysis on training cohort based on TP53 mutation variables, identified >0.5% as the best cutoff. In keeping with this cutoff, TP53 mutated patients experienced a significantly shorter OS than wt patients. Again cases with clonal and subclonal mutation experienced the same OS (Fig.I). Importantly, the cutoff found in the training cohort was able to reproduce the very same results also in the validation cohort both in term of mutation per se and in terms of clonal and subclonal TP53 mutations (Fig.J).
Conclusion. i) By applying ERIC recommendations and a rigorous pipeline of analysis, TP53mut impacted on OS also with VAF <1%; ii) del17p associated with short OS only when detectable in >10% of nuclei. These cutoffs may be employed for the clinical management of CLL patients.
Di Raimondo:Takeda: Consultancy; Amgen: Consultancy, Honoraria, Research Funding. Rossi:Gilead: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Abbvie: Honoraria, Other: Scientific advisory board; Janseen: Honoraria, Other: Scientific advisory board; Roche: Honoraria, Other: Scientific advisory board; Astra Zeneca: Honoraria, Other: Scientific advisory board.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal