In this issue of Blood, Berzuini et al1 describe immunoglobulin G (IgG) bound to the red blood cells (RBCs) of patients with COVID-19 and associate this bound IgG with increased RBC transfusion requirements. The intrigue behind these observations is not just the high (46%) direct antiglobulin test (DAT) positivity rate, but also the novel finding that eluates (ie, antibodies stripped from the surface of the reactive RBCs) from these DAT-positive patients react not with standard-reagent RBCs but exclusively with RBCs from DAT-negative COVID-19 patients.
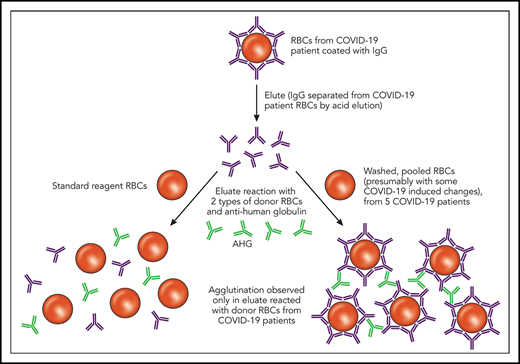
Elution studies are performed to separate bound Ig from a patient’s RBCs in the setting of an IgG-positive DAT; this separation is typically accomplished using pH or temperature changes. As depicted, the eluates from COVID-19 patients with an IgG-positive DAT did not show agglutination after incubation with commercially available reagent RBCs. However, the same eluates incubated with RBCs derived from other COVID-19 patients showed agglutination. Professional illustration by Patrick Lane, ScEYEnce Studios. AHG, anti-human globulin.
Elution studies are performed to separate bound Ig from a patient’s RBCs in the setting of an IgG-positive DAT; this separation is typically accomplished using pH or temperature changes. As depicted, the eluates from COVID-19 patients with an IgG-positive DAT did not show agglutination after incubation with commercially available reagent RBCs. However, the same eluates incubated with RBCs derived from other COVID-19 patients showed agglutination. Professional illustration by Patrick Lane, ScEYEnce Studios. AHG, anti-human globulin.
Although the association of DAT positivity and anemia is limited by study design, the serologic findings may be teaching us something important about modifications to RBCs that occur in COVID-19. The authors are to be congratulated on the rapid dissemination of these data and on the clever idea of using RBCs from COVID-19 patients as blood bank reagents in addition to standard, commercially available reagent RBCs.
First, a quick overview of the DAT, also known as the direct Coombs test. The DAT is designed to identify IgG or complement (C3) bound to a patient’s own RBCs. A DAT positive for IgG could be caused by several things,2 including autoimmunity, drugs, or intravenous immune globulin, among others, and not all positive DATs are clinically significant. Thus, the significance and specificity of a DAT can be further elucidated by comparing the results of an RBC antibody screen (indirect antiglobulin test that evaluates antibody in the plasma or sera) with the DAT and evaluating the results of the eluate. As Berzuini et al describe, IgG was detected in most of the positive DATs in hospitalized patients with COVID-19, and all of the IgG-positive DATs re-tested using a flow cytometric–based assay were positive for IgG, which helped to exclude nonspecific artifacts. Furthermore, C3d was detected in 12% of the positive DATs, either in combination with IgG or in isolation. The eluates from the IgG-positive DATs were negative against standard-reagent RBCs but uniformly positive with a panel of washed RBCs generated from 5 DAT-negative patients with COVID-19 (see figure). Possible causes for the unique pattern of eluate reactivity observed in patients with COVID-19 are RBC membrane modifications, complement effects, or drug effects.
Could downstream effects of severe acute respiratory syndrome coronavirus 2 (SARs-CoV-2) infection lead to a modified RBC membrane, resulting in the unique IgG binding DAT pattern described? One existing model of RBC membrane modification related to infection involves neuraminidase released by Streptococcus pneumoniae, which cleaves terminal N-acetylneuraminic acid (sialic acid) from glycoproteins and glycolipids. This exposes the Thomsen-Friedenreich crypt T antigen and, in effect, converts self RBCs to non-self RBCs. There is a debate regarding the exact role that these non-self RBCs play in the atypical hemolytic uremic syndrome pathophysiology, although impaired complement factor H binding to the desialylated RBC membranes and alternative pathway complement activation are thought to be involved.3
How might complement be related to the findings by Berzuini et al? Gao et al4 have recently described lung biopsy samples from patients with severe COVID-19 disease showing C3-fragment deposition; it is thought that the SARS-CoV-2 nucleocapsid (N) protein induces mannose-binding lectin-associated serine protease 2 activation, leading to cleavage of the complement protein C4 and downstream complement activation. The higher than expected C3d positivity reported by Berzuini et al is particularly intriguing in this backdrop, and one wonders if even more complement binding might have been detected if the authors had also used a C3b-specific reagent. In other settings, C3b is well known to serve as a direct ligand for complement receptors and Fc receptor engagement.5
Moreover, complement receptors on RBC surfaces are important in the clearance of immune complexes in infectious or inflammatory disease.6 It is possible, but purely speculative, that there may be alterations of complement receptors on the RBC surface of COVID-19 patients, contributing not only to the DAT positivity but also to the pattern of eluate reactivity. Future studies of the roles that complement and complement receptors may play in COVID-19, through the lectin pathway, the classical pathway, or the alternative pathway,7 are eagerly awaited.
Although Berzuini et al did not find a relationship between DAT positivity and any of the medications administered, the possibility of a drug effect cannot be fully discounted because an IgG-positive DAT with a negative eluate against standard-reagent RBCs may be drug associated until proven otherwise.8 In their article, it is unclear what drugs the 5 COVID-19–positive patients whose RBCs were used for eluate testing might have been taking. Repeating the eluate testing using RBCs from different COVID-19–positive donors who were not taking any drugs could be one way to investigate this possibility, although by definition such individuals would be less ill. Alternatively, the authors could incorporate select, commonly identified drugs into the eluate testing with standard-reagent RBCs to evaluate for a drug-dependent effect.
In sum, the Coombs test results and associated clinical data described by Berzuini et al in hospitalized patients with COVID-19 add to our growing understanding of hematologic manifestations of SARS-CoV-2. Given the time-sensitive nature of the COVID-19 pandemic, the data from the samples collected over a 1-week period are not exhaustive and the conclusions are not definitive. However, the described results are thought-provoking, and they join the findings of other rapidly written manuscripts in raising important questions to be investigated by the hematology and transfusion medicine communities in the years to come.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

Comments
The deleterious effect of SARS-CoV-2 on red blood cells
(1) Berzuini et al.1 described the high frequency of erythrocyte antibodies and the special binding behavior of these IgG antibodies to red blood cells (RBCs) in COVID-19 patients and controls. The Coombs test results suggest modifications of the RBC membranes during the disease.
(2) The first reports regarding the association between AB0 blood groups and the different risk of SARS-CoV-2 infection came from hospitals in China. A few months later, various countries all over the world confirmed the involvement of ABO blood groups in the risk pattern for COVID-19.2 Up until today the underlying mechanism is matter of debate.
SARS-CoV-2 uses the angiotensin-converting enzyme 2 as a receptor for cell entry. Beside the receptor-binding domain of SARS-CoV-2 known to bind the ACE2 receptor, Gallaher3 located a second receptor-binding domain on the outer surface of the N-terminal domain of the spike protein. This binding domain shows sequence- and structural homologies with the lima bean lectin. The lima bean lectin was used for blood typing since it is an anti-A agglutinin which precipitates best with blood group A1 substances, moderately with A2 substances, very poorly with B substances and not at all with blood group 0.4
Due to Gallaher´s hypothesis the additional receptor-binding domain of SARS-CoV-2 selectively binds the N-acetylgalactosamine of blood group A glycoside. Given the expression of ABO antigens in various human tissues 5 the virus spreading might be basically influenced. Other effects of the receptor-binding domain are purely speculative, although the idea suggests itself that the modifications of the RBC membranes are attributable to a lectin activity of this domain. Therefore, the assignment of the Coombs test findings to the ABO blood groups would be of great interest.
References
1 Berzuini A, Bianco C, Paccapelo C, et al. . Red cell–bound antibodies and transfusion requirements in hospitalized patients with COVID-19. Blood. 2020 Aug 6; 136(6): 766–768. [PMCID: PMC7414594] [PMID: 32559762]
2 Focosi D. Anti-A isohaemagglutinin titres and SARS-CoV-2 neutralization: implications for children and convalescent plasma selection. Br J Haematol. 2020 Aug; 190(3): e148-e150. Epub 2020 Jul 8. [PMID: 32516462] [PMCID: PMC7300571]
3 Gallaher W. Localized sequence identities between SARS-Cov-2 spike protein and lima bean agglutinin specific for ABO blood group A. https://virological.org/t/similarities-between-sars-cov-2-spike-protein-and-lima-bean-lectin-specific-for-abo-blood-group-a/518
4 Sikder SK, Kabat EA, Roberts DD and Goldstein IJ. Studies on the combining site of the blood group A-specific lima bean lectin. Carbohydrate Research. 1986; 151: 247-260. [PMID: 3768892]
5 Oriol P, Mollicone R, Coullin P, et al. . Genetic regulation of the expression of ABH and Lewis antigens in tissues. APMIS. 1992; 100; Supp 27: 29-38. [PMID: 1520528]
Conflict-of-interest disclosure: The authors declare no competing financial interests