Key Points
piRNA-30473 promotes DLBCL progression by regulating m6A RNA methylation in DLBCL.
piRNA-30473 improves the prognostic stratification and National Comprehensive Cancer Network-International Prognostic Index score of DLBCL.
Abstract
The initiation and progression of diffuse large B-cell lymphoma (DLBCL) is governed by genetic and epigenetic aberrations. As the most abundant eukaryotic messenger RNA (mRNA) modification, N6-methyladenosine (m6A) is known to influence various fundamental bioprocesses by regulating the target gene; however, the function of m6A modifications in DLBCL is unclear. PIWI-interacting RNAs (piRNAs) have been indicated to be epigenetic effectors in cancer. Here, we show that high expression of piRNA-30473 supports the aggressive phenotype of DLBCL, and piRNA-30473 depletion decreases proliferation and induces cell cycle arrest in DLBCL cells. In xenograft DLBCL models, piRNA-30473 inhibition reduces tumor growth. Moreover, piRNA-30473 is significantly associated with overall survival in a univariate analysis and is statistically significant after adjusting for the National Comprehensive Cancer Network-International Prognostic Index in the multivariate analysis. Additional studies demonstrate that piRNA-30473 exerts its oncogenic role through a mechanism involving the upregulation of WTAP, an m6A mRNA methylase, and thus enhances the global m6A level. Integrating transcriptome and m6A-sequencing analyses reveals that WTAP increases the expression of its critical target gene, hexokinase 2 (HK2), by enhancing the HK2 m6A level, thereby promoting the progression of DLBCL. Together, the piRNA-30473/WTAP/HK2 axis contributes to tumorigenesis by regulating m6A RNA methylation in DLBCL. Furthermore, by comprehensively analyzing our clinical data and data sets, we discover that the m6A regulatory genes piRNA-30473 and WTAP improve survival prediction in DLBCL patients. Our study highlights the functional importance of the m6A modification in DLBCL and might assist in the development of a prognostic stratification and therapeutic approach for DLBCL.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most frequent subtype of lymphoid malignancy and is heterogeneous in terms of clinical presentation, morphology, and biology.1 Standard therapies such as rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone cure two-thirds of patients, even those in advanced stages. Approximately 30∼40% of patients are still not cured, even when treated with the current therapies.2 To date, the clinical prognostic index, represented by the National Comprehensive Cancer Network-International Prognostic Index (NCCN-IPI), discriminated low- and high-risk subgroups.3 However, the NCCN-IPI score is not completely consistent with the clinical course of DLBCL patients, suggesting that heterogeneity is ubiquitous. Three molecular subtypes have been identified by gene expression profiling, including germinal center B-cell–like DLBCL, activated B-cell–like DLBCL, and unclassified cases (the “type 3” group), and activated B-cell–like DLBCL tends to have worse survival.4,5 Therefore, further research based on molecular oncogenic mechanisms is necessary for improving prognostic stratification and generating new therapeutic strategies.
As the most abundant chemical modification of eukaryotic messenger RNA (mRNA), N6-methyladenosine (m6A) is known to influence various fundamental bioprocesses by regulating target gene expression.6,7 m6A regulatory proteins are composed of the “erasers” FTO and ALKBH5, the “readers” YTHDFs and IGF2BPs, and the “writers” METTL3, METTL14, and WTAP.8-11 Alterations in the m6A level mediate cell apoptosis, proliferation, self-renewal, and development.12-15 However, the potential role of m6A methylation in DLBCL progression remains unknown.
PIWI-interacting RNAs (piRNAs) account for the majority of small noncoding RNAs.16 piRNA can guide PIWI proteins to silence transposable elements by regulating DNA methylation.17-19 Despite the usual supposition that piRNAs have important functions in germline development, recent evidence suggests that piRNAs also play key roles in tumorigenesis.20,21 piRNA-823 has been reported to be involved in the development of multiple myeloma and gastric cancer.22,23 However, the functional mechanisms of piRNAs in epitranscriptomic regulation in DLBCL require further investigation.
Therefore, we aimed to investigate the expression level of piRNA-30473 in DLBCL and its correlation with prognosis to further explore its role in epitranscriptomic regulation in DLBCL and its potential contribution to DLBCL pathogenesis.
Materials and methods
Study cohorts
Patients who were newly diagnosed with DLBCL gave informed consent and were enrolled in this study, and samples were taken. The detailed clinical characteristics of these patients are provided in supplemental Table 1 (available on the Blood Web site). These included 6 DLBCL patients whose paraffin-embedded tissues were profiled using a microarray and an independent cohort of 42 patients whose paraffin-embedded tissues were profiled using quantitative polymerase chain reaction (qPCR). All samples were obtained from The Second Affiliated Hospital of Soochow University (Suzhou, China). All patients provided written informed consent approving the use of their samples under Institutional Review Board approval.
Cell lines
SU-DHL-8 and Farage cells were cultured at 37°C in an atmosphere containing 5% CO2 and in RPMI-1640 medium (Hyclone) supplemented with 10% fetal bovine serum. 293T cells were cultured in high-glucose-formulated Dulbecco’s modified Eagle medium with 10% fetal bovine serum. Cell line authentication was performed by cell line characterization core using short tandem repeat profiling (Genetic Testing Biotechnology Corporation).
piRNA-30473 silencing by antagomir treatment
Cholesterol-conjugated antagomirs have been shown to be effective in delivering antagomir-mediated silencing in vivo.24 The 2′-OMe and phosphorothioate modification significantly enhances potency and resistance to degradation for various antagomirs.25 In this study, vivo-grade antagomirs were obtained from GenePharma and modified by cholesterol, 2′-OMe, and phosphorothioate. Modified antagomirs have been demonstrated great longevity and stability and enable effective antagomir uptake into cells and gene silencing in vivo.26-29 Chemically modified hsa-piRNA-30473 antagomir (antagomir-30473) was used to inhibit piRNA-30473 expression, and nontarget antagomir (antagomir-NC) was used as a negative control. The sequence of antagomir-30473 was as follows:
5′-GsCsUUCUACGAGGGUCCAUGUUGCAGGAGAUGUsGsCsUsChol-3′. Electroporation of antagomir was performed using the Neon Transfection System (Invitrogen).
Dual-luciferase reporter assay
To identify the putative binding site of piRNA-30473 at the WTAP 3′ untranslated region (UTR), bioinformatics analysis was performed using the miRanda algorithm (www.microrna.org). The wild-type (WT) and mutant (MUT) target sites at the WTAP 3′ UTR were amplified by PCR and then cloned into a pmirGLO vector. In addition, to test whether WTAP or IGF2BP2 might alter the expression of hexokinase 2 (HK2), we cloned the 5′ UTR, which contained an m6A methylation site, into a pGL-3 basic vector and cotransfected it with a pRL-cytomegalovirus vector containing the Renilla luciferase gene as a control. The 293T cells (1 × 105 cells/well) were seeded in 24-well plates and cotransfected with both the WT or MUT constructs with either specific inhibitors or negative control using Lipofectamine 3000 (Invitrogen). Cells were harvested at 48 hours, and the firefly luciferase activity was normalized to that of Renilla luciferase.
Plasmid construction
Full-length complementary DNA of WTAP was synthesized and subcloned into the EcoRI and BamHI sites of a Lenti-cytomegalovirus-puro vector, and the constructs were verified by DNA sequencing.
m6A dot blot
Poly(A) RNA was purified from total cellular RNA using the PolyATtract mRNA Isolation System (Promega). Poly(A) RNA was denatured at 65°C for 5 minutes and spotted onto a nylon membrane (GE Healthcare), followed by UV crosslinking at UV 254 nm, 0.12 J/cm2. After blocking in phosphate-buffered saline (PBS) with Tween 20 containing 5% bovine serum albumin for 1 hour, the membrane was incubated with an anti-m6A antibody (1:1000 diluted) and a 680RD goat anti-rabbit immunoglobulin G secondary antibody, and the membrane was visualized using an Odyssey Imaging System. Then, 500 ng poly(A) RNA was spotted on the membrane, which was stained with 0.02% methylene blue in 0.3 M sodium acetate (pH 5.2) for 1 hour, and washed with ribonuclease-free water for 1 hour.
m6A quantification
The change in the global m6A levels in mRNA was measured by an EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) (Epigentek) following the manufacturer’s protocol. Poly(A) RNA (200 ng) was used for sample analysis.
In vivo tumor xenograft model
The animal experiments were conducted according to the institutional guidelines for the use of laboratory animals and after permission was acquired from the local ethical committee for animal experimentation. Four-week-old female nonobese diabetic Prkdcscid Il2rgtm1/Bcgen (NSG) mice weighing ∼10 to 15 g were purchased from Biocytogen Company and fed in the specific pathogen–free barrier system. Mice were subcutaneously injected in the left flank with 106 SU-DH-L8 cells in 50% Matrigel (Corning). Tumor volume was monitored every other day using electronic digital calipers in 2 dimensions. Tumor volume was calculated using the following formula: tumor volume (mm3) = (smallest diameter2 × largest diameter)/2. When tumors were ∼100 mm3 ± 10%, the mice were randomly divided into 2 groups. One group was intraperitoneally injected with antagomir-30473 (16 mg/kg per day), and the other groups received PBS every other day consecutively for a total of 5 times.
Statistical analysis
In the preliminary study, a Mann-Whitney U test was performed to compare the expression of piRNAs between the favorable-prognosis group that received immunochemotherapy and achieved complete remission (overall survival [OS] >2 years) and the poor-prognosis group that received immunochemotherapy (OS <1 year). Then, we further analyzed the effect of piRNA expression on DLBCL patient outcomes.
The association between piRNAs and patient outcomes was analyzed via univariate and multivariate Cox regression models. We performed Benjamini-Hochberg correction for multiple comparisons. The OS was plotted and compared using the Kaplan-Meier method, and we separated the patients into 2 groups based on the median values for piRNA-30473 and WTAP expression. A Cox proportional hazards model was employed to compute the hazard ratios and accompanying 95% confidence intervals.
The area under the receiver-operating characteristic curve (AUC) was used to assess the predicted validity of the NCCN-IPI model and the NCCN-IPI + piRNA model. To avoid overfitting, we tested the performance of our model by means of 10-fold cross-validation.
All statistical analyses were performed in R (version 3.2.3; https://www.r-statistics.com/). A value of P < .05 was considered significant.
Other detailed assays are available in supplemental Methods.
Results
piRNA-30473 is elevated in DLBCL and is a negative prognostic factor for DLBCL patients
The identification of tissue-based biomarkers is critical for DLBCL research. In biomarker research, fresh frozen samples are more accurate, but in consideration of collecting many clinical samples with a long follow-up, formalin-fixed paraffin-embedded (FFPE) tissue samples with complete pathological and clinical data were also included.30 Moreover, in contrast to mRNA, small RNAs possess more stability in FFPE tissues because of their small size.31 Thus, we focused on FFPE DLBCL tissues to study the expression levels of small RNAs that may be associated with clinical outcome in patients with DLBCL. Microarray analysis was first performed to detect the expression levels of small RNAs in FFPE DLBCL tissues. By analyzing the microarray data, we identified 4 piRNAs in the favorable-prognosis group that were significantly downregulated or upregulated (>1.5-fold change; false discovery rate <0.05) compared with those in the poor-prognosis group (Figure 1A). Based on the microarray study, we concentrated on piRNAs that could be differentially expressed between DLBCL patients and could act as biomarkers for prognosis. Furthermore, we obtained paraffin-embedded tissues derived from 42 other DLBCL patients. The expression levels of these 4 piRNAs in patients with DLBCL were further confirmed using qPCR. We found that the piRNA-30473 expression level was significantly increased in patients with a poor prognosis relative to patients with a favorable prognosis. Conversely, no significant difference was found in the expression of the other 3 piRNAs in the 2 groups (Figure 1B).
piRNA-30473 is elevated in DLBCL and is a negative prognostic factor for patients with DLBCL. (A) Heat map representation of transcriptome array data on the expression levels of piRNAs in DLBCL patients with a poor prognosis (PP) (n = 3) and DLBCL patients with a favorable prognosis (FP) (n = 3). (B) qPCR analysis of piRNA expression in DLBCL patients with a poor prognosis (n = 21) and DLBCL patients with a favorable prognosis (n = 21). ns, not significant; **P < .01. (C) OS was plotted and compared using the Kaplan-Meier method and log-rank test. piRNA-30473 was dichotomized at the median based on low vs high expression.
piRNA-30473 is elevated in DLBCL and is a negative prognostic factor for patients with DLBCL. (A) Heat map representation of transcriptome array data on the expression levels of piRNAs in DLBCL patients with a poor prognosis (PP) (n = 3) and DLBCL patients with a favorable prognosis (FP) (n = 3). (B) qPCR analysis of piRNA expression in DLBCL patients with a poor prognosis (n = 21) and DLBCL patients with a favorable prognosis (n = 21). ns, not significant; **P < .01. (C) OS was plotted and compared using the Kaplan-Meier method and log-rank test. piRNA-30473 was dichotomized at the median based on low vs high expression.
The clinical characteristics of the patients are listed in supplemental Table 1. Kaplan-Meier analysis showed that high piRNA-30473 levels in patients with DLBCL were associated with poor OS (Figure 1C; supplemental Table 2). To further evaluate the value of piRNA-30473 as a prognostic signature in DLBCL, we incorporated the clinical information captured by the NCCN-IPI and piRNA-30473 signature into the multivariate model. Multivariate analysis indicated that the piRNA-30473 expression level was a statistically significant unfavorable prognostic factor (supplemental Table 2). Next, an AUC analysis with cross-validation was performed. The combination of the piRNA-30473 signature and NCCN-IPI had a better prediction of OS than without the piRNA-30473 signature (supplemental Table 3). Taken together, we found that the piRNA-30473 signature can be used as a biomarker of prognosis and developed the prognostic stratification of DLBCL patients.
piRNA-30473 inhibition decreases the proliferation and tumorigenesis of DLBCL
To further explore the functional role of piRNA-30473 in DLBCL, we depleted piRNA-30473 using antagomir-30473 in human DLBCL cells. Quantitative reverse transcription PCR was used to detect the efficiency of depletion after transfection (supplemental Figure 1A). We next found that depletion of piRNA-30473 in DLBCL cells resulted in reduced proliferation compared with the control (Figure 2A; supplemental Figure 1B). Moreover, cell cycle analysis showed that inhibition of piRNA-30473 in DLBCL cells enhanced the fraction of cells in G1 phase and diminished the fractions of cells in S phase and G2 phase (Figure 2B-C; supplemental Figure 1C-D). However, depletion of piRNA-30473 did not induce apoptosis (Figure 2D; supplemental Figure 1E).
piRNA-30473 inhibition decreases the tumorigenesis of DLBCL. (A) SU-DHL-8 cells (antagomir-NC or antagomir-30473 transfected) were harvested at 24, 48, and 72 hours after transfection. Cell proliferation was assessed by CCK-8 assays. (B-C) Cell cycle distribution of control and piRNA-30473-depleted SU-DHL-8 cells indicated by DNA content index. (D) Percentage of apoptotic cells in control and piRNA-30473-depleted SU-DHL-8 cells. (E) Schematic illustration of the DLBCL mouse model treated with antagomir-30473 or PBS in vivo injection. (F) DLBCL mice were treated with antagomir-30473 or PBS. Tumor volumes were monitored every other day. Error bars represent the mean ± standard deviation (SD) of 5 independent experiments. ***P < .001; ****P < .0001.
piRNA-30473 inhibition decreases the tumorigenesis of DLBCL. (A) SU-DHL-8 cells (antagomir-NC or antagomir-30473 transfected) were harvested at 24, 48, and 72 hours after transfection. Cell proliferation was assessed by CCK-8 assays. (B-C) Cell cycle distribution of control and piRNA-30473-depleted SU-DHL-8 cells indicated by DNA content index. (D) Percentage of apoptotic cells in control and piRNA-30473-depleted SU-DHL-8 cells. (E) Schematic illustration of the DLBCL mouse model treated with antagomir-30473 or PBS in vivo injection. (F) DLBCL mice were treated with antagomir-30473 or PBS. Tumor volumes were monitored every other day. Error bars represent the mean ± standard deviation (SD) of 5 independent experiments. ***P < .001; ****P < .0001.
In addition, SU-DHL-8 xenograft DLBCL models were generated to assess the effect of piRNA-30473 on DLBCL progression in vivo (Figure 2E). Treatment with antagomir-30473 resulted in a significant decrease in tumor volume relative to that of the control, with an average tumor size of 1307.41 ± 264.32 mm3 vs 306.88 ± 149.46 mm3 (Figure 2F). These data indicated that silencing piRNA-30473 could inhibit DLBCL cell activity in vitro and in vivo.
piRNA-30473 mediates m6A methylation by regulating the expression of WTAP
Dysregulated DNA methylation induced by piRNAs is a hallmark of cancer,22,32 whereas the function of piRNAs in m6A RNA modification remains unclear. To further explore the role of piRNA-30473 in epitranscriptomic regulation in DLBCL, the global m6A methylation level was detected at 48 hours after antagomir-30473 transfection. Notably, the global m6A level in antagomir-30473-transfected cells was decreased compared with that in the control cells (Figure 3A-B; supplemental Figure 2A-B). Methyltransferase or demethylase primarily modify the m6A level.8-11 We hypothesized that the dysregulation of the key m6A methyltransferases or demethylases may contribute to the abnormal m6A modification profile in DLBCL. To investigate whether piRNA-30473 mediated the m6A level indirectly by regulating RNA methyltransferases or RNA demethylases, we then examined the levels of METTL3, METTL14, WTAP, FTO, and ALKBH5 at 48 hours after antagomir-30473 transfection. We found that silencing piRNA-30473 reduced WTAP expression at the mRNA and protein levels (Figure 3C; supplemental Figure 2C-D). However, the expression of other proteins was not significantly different between the 2 groups. In addition, immunohistochemistry showed that treatment with antagomir-30473 partly reduced WTAP expression in DLBCL xenograft models (Figure 3D).
piRNA-30473 mediates m6A methylation by regulating the expression of WTAP. (A-B) m6A levels of SU-DHL-8 cells with or without piRNA-30473 knockdown were detected using the EpiQuik m6A RNA Methylation Quantification Kit (A) and m6A dot blot assays (B). MB, methylene blue staining (as loading control). (C) SU-DHL-8 was transiently transfected with antagomir-30473 or antagomir-NC. qPCR was performed to determine METTL3, METTL14, WTAP, ALKBH5, and FTO mRNA levels. (D) Representative immunohistochemical staining of WTAP in tumor xenografts is shown (original magnification ×40). WTAP-positive particles (brown) are apparent in the nucleus (nuclei stained with hematoxylin, blue). Scale bars, 100 μm. (E) Cell lysates were subjected to coimmunoprecipitation (IP) with anti-WTAP antibody or rabbit immunoglobulin G (IgG) as a control, followed by immunoblotting with the respective antibodies, as shown. (F) A schematic representation of the interaction between piRNA-30473 and the 3′ UTR of WTAP. (G) Dual-luciferase reporter assays showed the effect of piRNA-30473 on WTAP reporters with either WT or MUT binding sites. (H) SU-DHL-8 or Farage cells were transiently transfected with antagomir-30473 or antagomir-NC. The half-life of WTAP mRNA was measured. Error bars represent the mean ± SD of 3 independent experiments. ***P < .001.
piRNA-30473 mediates m6A methylation by regulating the expression of WTAP. (A-B) m6A levels of SU-DHL-8 cells with or without piRNA-30473 knockdown were detected using the EpiQuik m6A RNA Methylation Quantification Kit (A) and m6A dot blot assays (B). MB, methylene blue staining (as loading control). (C) SU-DHL-8 was transiently transfected with antagomir-30473 or antagomir-NC. qPCR was performed to determine METTL3, METTL14, WTAP, ALKBH5, and FTO mRNA levels. (D) Representative immunohistochemical staining of WTAP in tumor xenografts is shown (original magnification ×40). WTAP-positive particles (brown) are apparent in the nucleus (nuclei stained with hematoxylin, blue). Scale bars, 100 μm. (E) Cell lysates were subjected to coimmunoprecipitation (IP) with anti-WTAP antibody or rabbit immunoglobulin G (IgG) as a control, followed by immunoblotting with the respective antibodies, as shown. (F) A schematic representation of the interaction between piRNA-30473 and the 3′ UTR of WTAP. (G) Dual-luciferase reporter assays showed the effect of piRNA-30473 on WTAP reporters with either WT or MUT binding sites. (H) SU-DHL-8 or Farage cells were transiently transfected with antagomir-30473 or antagomir-NC. The half-life of WTAP mRNA was measured. Error bars represent the mean ± SD of 3 independent experiments. ***P < .001.
Previous studies suggested that WTAP mediates the recruitment of METTL3 and METTL14 to mRNA targets. In the absence of WTAP, the RNA-binding capability of METTL3 is strongly reduced.10 Therefore, we hypothesized that piRNA-30473 might regulate m6A methylation by reducing the binding of WTAP to METTL3 and METTL14. As expected, coimmunoprecipitation indicated that antagomir-30743 treatment decreased the association of WTAP with METTL3 and METTL14 (Figure 3E).
Previous research has shown that piRNAs exhibit imperfect complementarity with their mRNA targets.33,34 The theory of physical interaction between piRNAs and target genes prompted us to examine the cellular distribution of piRNA-30473. qPCR of nuclear and cytoplasmic fractions and RNA fluorescence in situ hybridization (supplemental Figure 2E-F) suggested that piRNA-30473 was located in both the nucleus and cytoplasm. Thus, we next determined whether piRNA-30473 had a putative binding site within WTAP mRNA using miRanda, a microRNA-specific target detection algorithm (Figure 3F). We found that piRNA-30473 had 1 binding site within the 3′ UTR of WTAP. To further verify that WTAP was a direct target gene of piRNA-30473, a dual-luciferase reporter assay was performed (supplemental Figure 2G), which showed that piRNA-30473 silencing caused a significant decrease in the luciferase activity of the reporter construct carrying the WT WTAP 3′ UTR relative to the control. This decrease was abrogated when the putative binding site was mutated (Figure 3G). Moreover, we found that knockdown of piRNA-30473 resulted in decreased stability of WTAP mRNA (Figure 3H). Therefore, piRNA-30473 reduced WTAP mRNA decay and then enhanced mRNA stability by binding to the 3′ UTR of WTAP.
Based on these observations, we concluded that piRNA-30473 was required for maintaining the expression and function of WTAP in DLBCL.
WTAP plays an oncogenic role in DLBCL
We next investigated whether WTAP played a crucial role in the clinical outcome of DLBCL, and immunohistochemical analysis was used to assess WTAP protein expression in DLBCL patient samples. Immunohistochemical analysis showed that WTAP was highly expressed in patients with a poor prognosis (Figure 4A). Moreover, gene expression profiling of DLBCL patients was analyzed using the Gene Expression Omnibus (GEO) database (GSE10846), and the analysis results indicated that the elevated expression of WTAP predicted a poor prognosis (Figure 4B; supplemental Table 4). However, the AUC analysis showed that the combination of the WTAP signature and NCCN-IPI failed to improve the prognostic efficacy of the NCCN-IPI (supplemental Table 5).
WTAP is a functionally important target gene of piRNA-30473 in DLBCL. (A) Representative immunohistochemical staining (original magnification ×40) of WTAP in DLBCL patients is shown. WTAP-positive particles (brown) are apparent in the nucleus (nuclei stained with hematoxylin, blue). Original magnification ×40. (B) OS was plotted and compared using the Kaplan-Meier method and log-rank test. WTAP was dichotomized at the median based on low vs high expression. (C) SU-DHL-8 cells (si-NC or si-WTAP-transfected) were harvested at 24, 48, and 72 hours after transfection. Cell proliferation was assessed by CCK-8 assays. (D-E) Cell cycle distribution of control and WTAP-depleted SU-DHL-8 cells examined by DNA content index at 48 hours after transfection. (F) The percentage of apoptotic cells in control and WTAP-depleted SU-DHL-8 cells was examined at 48 hours after transfection. Error bars represent the mean ± SD of 3 independent experiments. ****P < .0001.
WTAP is a functionally important target gene of piRNA-30473 in DLBCL. (A) Representative immunohistochemical staining (original magnification ×40) of WTAP in DLBCL patients is shown. WTAP-positive particles (brown) are apparent in the nucleus (nuclei stained with hematoxylin, blue). Original magnification ×40. (B) OS was plotted and compared using the Kaplan-Meier method and log-rank test. WTAP was dichotomized at the median based on low vs high expression. (C) SU-DHL-8 cells (si-NC or si-WTAP-transfected) were harvested at 24, 48, and 72 hours after transfection. Cell proliferation was assessed by CCK-8 assays. (D-E) Cell cycle distribution of control and WTAP-depleted SU-DHL-8 cells examined by DNA content index at 48 hours after transfection. (F) The percentage of apoptotic cells in control and WTAP-depleted SU-DHL-8 cells was examined at 48 hours after transfection. Error bars represent the mean ± SD of 3 independent experiments. ****P < .0001.
To gain insight into the mechanism by which WTAP controls DLBCL development and prognosis, we inhibited WTAP expression by transfecting WTAP-specific small interfering RNA (siRNA) into SU-DHL-8 and Farage cells, which are derived from DLBCL. Similar effects of WTAP depletion and piRNA-30473 depletion were observed in DLBCL cells. The global m6A level was decreased in WTAP-depleted cells (supplemental Figure 3A-C). In addition, we found that depletion of WTAP also induced growth inhibition and cell cycle arrest (Figure 4C-E; supplemental Figure 3D-F) without evidence of apoptosis (Figure 4F; supplemental Figure 3G). We sought to determine whether restoring the expression of WTAP in cells with downregulated piRNA-30473 could reverse the altered biological activity. These results of the rescue experiment indicated that the effects of piRNA-30473 inhibition could be largely rescued by overexpression of WTAP (supplemental Figure 4). Collectively, our data demonstrated that WTAP was a functionally important target of piRNA-30473 and played an oncogenic role in DLBCL.
Identification of WTAP targets by RNA-seq and m6A-seq
To further validate the effects of WTAP-mediated m6A methylation on DLBCL cells, m6A sequencing (m6A-seq) was performed on control and WTAP-deficient SU-DHL-8 cells. Consistent with published studies, the consensus motif DRACH (D = A, G, or U; R = A or G; H = A, U, or C) was enriched in the immune-purified RNA (Figure 5A). Moreover, 23 to 29 million reads were generated from each m6A-seq or RNA sequencing (RNA-seq) (also serving as the input control of the corresponding m6A-seq) library.
Identification of WTAP target genes by high-throughput RNA-seq and m6A-seq. (A) The consensus motif was identified by HOMER with m6A-seq peaks in SU-DHL-8 cells with or without WTAP knockdown. (B) Graphs of m6A peak distribution showing the proportion of total m6A peaks in the indicated regions in control and WTAP-deficient cells. CDS, coding sequence; TSS, transcription start site. (C) Distribution of transcripts with a significant change in both m6A level and expression level in WTAP knockdown compared with control SU-DHL-8 cells. (D) Gene set enrichment analysis plots for WTAP-regulated genes, as determined by RNA-seq. (E) The m6A abundances in HK2 mRNA transcripts in WTAP knockdown and control SU-DHL-8 cells. (F) Relative mRNA levels of HK2 from qPCR analysis in WTAP knockdown compared with control DLBCL cells. (G) Gene-specific m6A qPCR validation of m6A level changes in HK2 in WTAP knockdown compared with control DLBCL cells. (H) Dual-luciferase reporter assays showed the effect of WTAP on HK2 reporters with either WT or MUT binding sites. (I) Schematic diagram of knocking out the m6A RNA methylation site on the 5′ UTR of HK2 using the CRISPR-Cas9 genome editing method. (J) Farage-knockout (Farage-KO) cells (si-NC or si-WTAP-transfected) were harvested at 24, 48, and 72 hours after transfection. Cell proliferation was assessed by CCK-8 assays. Error bars represent the mean ± SD of 3 independent experiments. **P < .01; ****P < .0001.
Identification of WTAP target genes by high-throughput RNA-seq and m6A-seq. (A) The consensus motif was identified by HOMER with m6A-seq peaks in SU-DHL-8 cells with or without WTAP knockdown. (B) Graphs of m6A peak distribution showing the proportion of total m6A peaks in the indicated regions in control and WTAP-deficient cells. CDS, coding sequence; TSS, transcription start site. (C) Distribution of transcripts with a significant change in both m6A level and expression level in WTAP knockdown compared with control SU-DHL-8 cells. (D) Gene set enrichment analysis plots for WTAP-regulated genes, as determined by RNA-seq. (E) The m6A abundances in HK2 mRNA transcripts in WTAP knockdown and control SU-DHL-8 cells. (F) Relative mRNA levels of HK2 from qPCR analysis in WTAP knockdown compared with control DLBCL cells. (G) Gene-specific m6A qPCR validation of m6A level changes in HK2 in WTAP knockdown compared with control DLBCL cells. (H) Dual-luciferase reporter assays showed the effect of WTAP on HK2 reporters with either WT or MUT binding sites. (I) Schematic diagram of knocking out the m6A RNA methylation site on the 5′ UTR of HK2 using the CRISPR-Cas9 genome editing method. (J) Farage-knockout (Farage-KO) cells (si-NC or si-WTAP-transfected) were harvested at 24, 48, and 72 hours after transfection. Cell proliferation was assessed by CCK-8 assays. Error bars represent the mean ± SD of 3 independent experiments. **P < .01; ****P < .0001.
m6A-seq identified 17 274 and 1900 m6A peaks from 7560 to 1007 m6A-modified transcripts in control and WTAP-deficient cells, respectively. In total, 5701 peaks (3734 transcripts) showed an increase in m6A levels, whereas 5702 peaks (2119 transcripts) presented a decrease m6A methylation in WTAP knockdown cells relative to control cells. In line with previous analysis, m6A sites were primarily found in both exons and 3′ UTRs, with the highest enrichment of m6A residues located near the stop codon (Figure 5B). Moreover, RNA-seq revealed that 186 transcripts were significantly downregulated, while 318 transcripts were significantly upregulated. Since WTAP is an m6A methyltransferase, transcripts bearing hypomethylated m6A peaks consequent to WTAP depletion are likely to be targets of WTAP. Filtering the hypomethylated m6A peaks that occurred in differentially expressed genes resulted in the identification of 136 genes (Figure 5C), as listed in supplemental Table 6.
In addition, we examined the correlation between WTAP and the above genes in DLBCL patient samples (GSE10846, GSE31312, GSE32918, GSE38202, GSE56313, and GSE57611), and we found that the expression of 28 genes was correlated with WTAP in DLBCL (|R|> 0.1, P < .05). Among these potential target genes, only HK2 had a significant correlation with WTAP in all data sets (supplemental Figure 5A). HK2, which encodes the enzyme hexokinase 2, contributes to the high glycolytic rate in tumors.35 On the other hand, HK2 is required for promoting growth during hypoxic stress in DLBCL.36 Moreover, gene set enrichment analysis indicated that potential target genes of WTAP were significantly correlated with hypoxia signatures (the gene sets ELVIDGE_HIF1A_AND_HIF2A_TARGETS_DN, ELVIDGE_HYPOXIA_UP, and MENSE_HYPOXIA_UP) (Figure 5D), suggesting that HK2 was a critical target gene of WTAP in DLBCL. To further study whether HK2 could predict poor clinical outcome in DLBCL patients, the GEO database (GSE10846) was used. Kaplan-Meier analysis showed that high HK2 levels in patients with DLBCL were associated with poor OS (supplemental Figure 5B), and the addition of HK2 to the NCCN-IPI could improve the prognostic stratification of patients with DLBCL (supplemental Tables 7 and 8).
Our m6A-seq data indicated that WTAP targeted the 5′ UTR of HK2 transcripts, and WTAP knockdown caused a significant decrease in the m6A level of the 5′ UTR (Figure 5E). To validate the m6A-seq and RNA-seq results, we next conducted gene-specific m6A qPCR and qPCR assays for HK2 in WTAP-deficient cells (Figure 5F-G). Luciferase reporter vectors (pGL3-HK2-WT or pGL3-HK2-MUT) were constructed to further demonstrate the reliability of m6A-seq data (supplemental Figure 5B). As expected, WTAP knockdown substantially reduced the luciferase activity of the pGL3-HK2-WT vector, which had intact m6A sites, while mutations in the m6A sites abrogated the inhibition (Figure 5H). Furthermore, to investigate whether HK2 was the unique target of WTAP, we deleted the WTAP binding site on the HK2 5′ UTR using a CRISPR-Cas9 strategy in Farage (supplemental Figure 5D). Loss of the m6A RNA methylation site on HK2 reversed the WTAP-mediated phenotypic changes, confirming the mechanistic role of the WTAP/HK2 axis in DLBCL (Figure 5I-J). The above results indicated that HK2 is a direct downstream target of WTAP in DLBCL.
The m6A RNA reader IGF2BP2 is essential for the posttranscriptional regulation of HK2
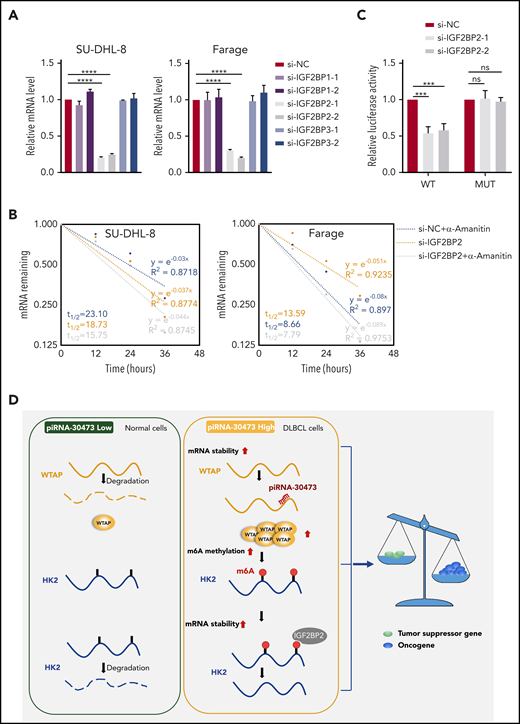
Our results suggested that alternative m6A reading processes might exist that recognize m6A sites in WTAP target transcripts to promote their stability. IGF2BPs are well-established m6A readers, and each recognizes thousands of methylated transcripts in mammalian cells.37 All 3 IGF2BPs preferentially bind to the “GGAC” consensus sequence m6A core motif, and our m6A-seq results indicated that the IGF2BP-binding site was located in the 5′ UTR of HK2. To determine whether the expression of HK2 was affected by IGF2BPs, we detected the expression levels of HK2 in IGF2BP knockdown cells. qPCR demonstrated that the transcript levels of HK2 were reduced significantly in IGF2BP2-deficient cells (Figure 6A). As expected, the mRNA stability experiment revealed that HK2 tended to be less stable in IGF2BP2-deficient cells (Figure 6B). Moreover, in the GEO data sets, we found that the expression of HK2 was correlated with IGF2BP2 (supplemental Table 9).
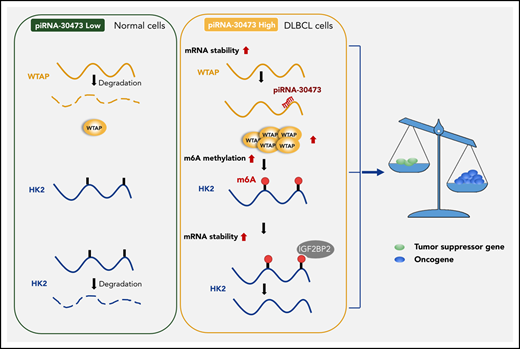
WTAP-mediated regulation of HK2 depends on its m6A methylase activity. (A) Relative mRNA levels of HK2 from qPCR analysis in IGF2BP knockdown cells compared with control cells. (B) SU-DHL-8 or Farage cells were transiently transfected with IGF2BP2-siRNA or siRNA control. The half-life of HK2 mRNA was measured. (C) Dual-luciferase reporter assays showed the effect of IGF2BP2 on HK2 reporters with either WT or MUT binding sites. (D) Schematic diagram depicting the mechanism by which piRNA-30473 contributes to tumorigenesis and poor prognosis in DLBCL by regulating m6A RNA methylation and thereby triggering corresponding signaling cascades. Error bars represent the mean ± SD of 3 independent experiments. ***P < .001; ****P < .0001.
WTAP-mediated regulation of HK2 depends on its m6A methylase activity. (A) Relative mRNA levels of HK2 from qPCR analysis in IGF2BP knockdown cells compared with control cells. (B) SU-DHL-8 or Farage cells were transiently transfected with IGF2BP2-siRNA or siRNA control. The half-life of HK2 mRNA was measured. (C) Dual-luciferase reporter assays showed the effect of IGF2BP2 on HK2 reporters with either WT or MUT binding sites. (D) Schematic diagram depicting the mechanism by which piRNA-30473 contributes to tumorigenesis and poor prognosis in DLBCL by regulating m6A RNA methylation and thereby triggering corresponding signaling cascades. Error bars represent the mean ± SD of 3 independent experiments. ***P < .001; ****P < .0001.
Furthermore, the pGL3-HK2-WT and pGL3-HK2-MUT vectors were used to establish that IGF2BP2 targeted the 5′ UTR of HK2 mRNA (supplemental Figure 5C). The luciferase activity of the pGL3-HK2-WT vector was reduced through inhibition of IGF2BP2, whereas this effect disappeared with the pGL3-HK2-MUT vector (Figure 6C). Thus, the RNA-binding protein IGF2BP2 functioned collectively with WTAP to affect HK2 mRNA stability and expression in DLBCL cells.
Discussion
DLBCL is considerably heterogeneous lymphoid malignancy with diverse morphologies and molecular features.38 Therefore, clarifying the molecular mechanism of DLBCL tumorigenesis and progression has important value in basic theory and clinical applications. Recently, the molecular mechanisms of piRNA in tumorigenesis have been found in various tumor types.21,23 Here, we aimed to identify a piRNA that may serve as an independent biomarker for the tumorigenesis and progression of DLBCL. Our findings add a biomarker, piRNA-30473, into the catalog of prognostic factors in DLBCL (Figure 1) and reveal its roles in DLBCL cell proliferation and cell cycle progression (Figure 2; supplemental Figure 1).
More recently, piRNA-823 has been reported to be involved in the tumorigenesis of multiple myeloma and participate in regulating de novo DNA methylation and the tumor microenvironment.22 However, the underlying mechanisms of piRNA action on aberrant RNA methylation remain to be fully elucidated. m6A is the most abundant epigenetic mark on eukaryotic mRNAs among the >100 types of chemical modifications on RNA. Therefore, we clarified a link between altered piRNA-30473 and aberrant m6A methylation in DLBCL. piRNA-30473 inhibition led to a marked reduction in WTAP (an m6A methyltransferase) expression at both the mRNA and protein levels, which was accompanied by significantly decreased m6A levels (Figure 3; supplemental Figure 2). Further analyses showed that piRNA-30473 enhanced the stability of WTAP by targeting a complementary sequence in the 3′ UTR. (Figure 4).
m6A modification plays a vital role in regulating a range of biological processes, including primary microRNA processing,39 alternative splicing,40 stem cell regeneration,41 and mRNA translation.37 In addition, the most recent research indicated that dysregulation of m6A modification is associated with leukemia.14,42,43 Although there is evidence that m6A may serve as a key regulator in leukemia, few studies have been performed in other hematological malignancies. To our knowledge, no investigation of its presence in DLBCL has been performed.
We therefore sought to address whether aberrant m6A methylation was related to disease progression and prognosis. In this study, we found that enhanced expression of WTAP was associated with poor prognosis in DLBCL patients. Consistent with the effects of piRNA-30473, WTAP actually displayed strong carcinogenic activity, maintaining DLBCL cell viability and normal cell cycle (Figure 4). The role of WTAP has been described in glioblastoma44 and acute myeloid leukemia.45 In the present study, we found for the first time that WTAP was overexpressed and played an oncogenic role through its methylation activity in DLBCL (Figure 4).
We next investigated the role of the m6A modification and addressed the mechanisms by which WTAP participated in the biology of DLBCL. Our m6A-seq and subsequent validation suggested that HK2 was the critical target gene of WTAP. Inhibition of WTAP reduced the m6A level of HK2, which led to the downregulation of HK2 at the RNA level (Figure 5). HK2, an essential kinase of glucose metabolism,46 is associated with tumor cell proliferation via enhanced aerobic glycolysis.35,47,48 It is worth mentioning that research on hypoxic control in the progression of DLBCL provides strong support for the direct contribution of HK2 in B-cell lymphoma development and suggests that HK2 is a key metabolic driver of the DLBCL phenotype.36 Based on the above studies, we hypothesized that the piRNA-30473/WTAP/HK2 axis may play a critical role in the pathogenesis of DLBCL.
Previous studies suggest that mRNA transcripts with m6A modifications tend to be more stable, largely due to regulation by m6A readers such as IGF2BPs.29,37 Furthermore, the selective binding of IGF2BPs to m6A-modified HK2 has been confirmed in HepG27 and GM12878 cells.29 Moreover, we found that HK2 and IGF2BP2 exhibited a significantly positive correlation in mRNA expression across all 6 data sets. We next showed that depleted expression of IGF2BP2 substantially reduced HK2 expression by shortening the transcript’s half-life (Figure 6).
In summary, we provide compelling in vitro and in vivo evidence to demonstrate that piRNA-30473 contributes to tumorigenesis and poor prognosis of DLBCL by regulating m6A RNA methylation, thereby triggering downstream signaling cascades. Our study highlights the functional importance of the m6A modification machinery in DLBCL and provides profound insights into the epigenetic mechanisms underlying tumorigenesis by revealing a previously undiscovered mechanism of gene regulation in DLBCL. In addition, given the functional importance of piRNA-30473 in tumorigenesis and disease progression, targeting the piRNA-30473/WTAP/HK2 axis with selective inhibitors could represent a promising therapeutic strategy for treating DLBCL (Figure 6D).
All information related to microarray analysis, RNA-seq, and m6A-seq has been deposited to the GEO under accession numbers GSE128852 (microarray analysis) and GSE129469 (RNA-seq and m6A-seq).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported, in part, by the Science and Technology Development Project of Suzhou City (SS201856), the National Clinical Research Center for Hematologic Diseases, the National Natural Science Foundation of China (81670191 and 81673448), Jiangsu Social Development Project New Clinical Diagnosis and Treatment Technology (SBE2019740362), Clinical Key Diagnosis and Treatment Technologies in Suzhou City (LCZX201805), and The Fifth Phase of “333 High-level Talents Training Project” in Jiangsu Province (BRA2018138).
Authorship
Contribution: H.H., G.F., S.S., Y.J., W. Zhang, Q.S., and X.X. designed the research; H.H. and G.F. wrote the paper; C.Q. and G.F. analyzed the data; and B.L. and W. Zhuang contributed vital new reagents or analytical tools.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bingzong Li, Department of Haematology, The Second Affiliated Hospital of Soochow University, San Xiang Rd 1055, Suzhou 215006, China; e-mail: lbzwz0907@hotmail.com; and Wenzhuo Zhuang, Department of Cell Biology. School of Biology & Basic Medical Sciences, Soochow University, Ren Ai Rd 199, Suzhou 215123, China; e-mail: zhuangwenzhuo@suda.edu.cn.
REFERENCES
Author notes
H.H. and G.F. contributed equally to this study.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal