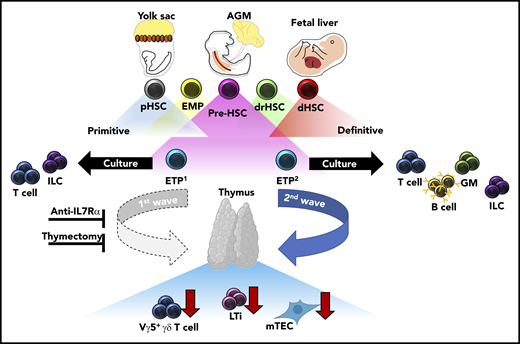
In this issue of Blood, Elsaid et al reinforce the temporal and spatial complexity of immune layering during embryogenesis by intricately dissecting 2 distinct waves of early thymic progenitors (ETPs) that differentially contribute to normal thymic organogenesis and homeostasis.1 The concept of “layered immunity,” in the form of waves of different immune cells derived from developmentally distinct progenitor populations, was initially described for innate-like lymphocytes, including B1-B cells and γ-δ T cells.2,3 Cellular compartments generated by layered immunity now extend beyond subsets of innate-like B and T cells to include tissue-resident macrophages,4 innate lymphoid cells (ILC),5 and mast cells.6
During fetal development, transient waves of progenitors arise and generate distinct immune cells that persist and contribute to the mature immune system. Elsaid et al elegantly demonstrate that 2 distinct waves of ETPs arise from HSC-derived progenitors and differentially contribute to thymic innate-immune cell establishment and mTEC maturation. Blockade of the first, but not the second, wave of ETPs by anti-IL7Rα injection results in significantly less thymic LTi cells, Vγ5+ γδ T cells, and mature mTECs. These results were recapitulated in neonates after complete thymectomy. Altogether, these data suggest that the initial wave of ETPs specifically contributes to the establishment of innate-immune cell compartments in the developing thymus and is required for the maturation of AIRE-expressing mTECs. AGM, aorta-gonad-mesonephros; dHSC, definitive HSC; drHSC, developmentally-restricted HSC; EMP, erythro-myeloid progenitor; GM, granulocyte/monocyte; pHSC, primitive HSC.
During fetal development, transient waves of progenitors arise and generate distinct immune cells that persist and contribute to the mature immune system. Elsaid et al elegantly demonstrate that 2 distinct waves of ETPs arise from HSC-derived progenitors and differentially contribute to thymic innate-immune cell establishment and mTEC maturation. Blockade of the first, but not the second, wave of ETPs by anti-IL7Rα injection results in significantly less thymic LTi cells, Vγ5+ γδ T cells, and mature mTECs. These results were recapitulated in neonates after complete thymectomy. Altogether, these data suggest that the initial wave of ETPs specifically contributes to the establishment of innate-immune cell compartments in the developing thymus and is required for the maturation of AIRE-expressing mTECs. AGM, aorta-gonad-mesonephros; dHSC, definitive HSC; drHSC, developmentally-restricted HSC; EMP, erythro-myeloid progenitor; GM, granulocyte/monocyte; pHSC, primitive HSC.
One of the outstanding questions regarding layered immunity is defining the unique interactions between waves of developmentally regulated immune cells and the developing tissues they seed. Importantly, these interactions serve to define both tissue-resident immune cell identity and function, and overall tissue homeostasis. When these interactions are dysregulated, they can lead to immune and tissue dysfunction. The authors contribute to this topic by defining differences in differentiation potential and innate-immune cell establishment between the first (embryonic day 13 [E13]) and second (E18) wave of ETPs in the developing thymus. Single-cell culture assays demonstrated restricted differentiation potential between E13 ETPs as compared to E18 ETPs, the former generating only lymphoid tissue inducer (LTi) cells and T cells. Furthermore, in vitro fetal thymic organ culture experiments, recapitulated in vivo, revealed LTi and Vγ5+ γδ T-cell generation was specifically attributed to E13 ETPs and not E18 ETPs, highlighting that thymic innate-immune cell generation is temporally regulated. In agreement with their limited differentiation potential, single-cell transcriptional analysis revealed that E13 ETPs appear to be transcriptionally primed toward LTi and invariant T-cell profiles. Importantly, these data suggest a previously unappreciated source of LTi cell generation outside of the fetal liver (FL), which is currently understood to be the primary source of LTi cells during fetal development. Furthermore, dependency of thymic tissue-resident Vγ5+γδ T cells, Vγ6+ γδ T cells in the lymph nodes, and medullary thymic epithelial cell (mTEC) maturation within the thymus was shown to be primarily attributed to E13 ETPs through experiments in which the first wave of thymic development was blocked with either a monoclonal anti-IL7Rα antibody or neonatal thymectomy. Altogether, these data strongly suggest that a functional thymus depends on temporally regulated layering from ETPs with unique differentiation potentials (see figure).
It is noteworthy that in their attempt to dissect the earliest origins of ETP, the authors used multiple lineage-tracing approaches to test the origin of different lineages. Using an established Il7raCreRosa26YFP lineage tracing model to detect the earliest progenitor of IL7R+ ETPs, the authors demonstrated that although YFP+ expressing cells marked by IL7R expression can be detected in the yolk sac (YS) as early as E9.5, as previously reported,7 these IL7R-marked progenitors lacked lymphoid potential in ex vivo culture. This is in stark contrast to the same (IL7R+) YS Kit+ progenitors at E10.5, which readily generated T-cell subsets under the same conditions. Interestingly, transcriptional profiling of E9.5 YFP+ and YFP− cells indicated that these 2 populations did not significantly differ in transcript expression, and only 6 of 47 YFP+ cells coexpressed IL7Rα transcripts. Transient expression of IL7ra message and surface expression was recently described in tissue-resident macrophages,8 suggesting a decoupling of conventional lymphoid-associated lineage commitment and differentiation potential during fetal hematopoiesis. Importantly, the author’s finding of transient expression of a lymphoid-associated lineage marker without accompanying lymphoid potential challenges our understanding of differentiation potential and lineage commitment, emphasizing the importance not only of transcriptional profiling but also of in vivo testing of lineage contribution.
The authors appropriately acknowledged that the constitutive Il7rαCreRosa26YFP lineage tracing model lacks the ability to faithfully trace the origin of embryonic-derived lymphoid cells due to YFP expression in downstream progenitor and mature cells. To address this point, they also employed a Csf1rMeriCreMerRosa26YFP lineage tracing model to better define the origin of the first wave of ETPs during embryogenesis. As ETPs were only labeled upon 4-hydroxytamoxifen pulse at E10.5 coincident with the emergence of the hematopoietic stem cells (HSCs) and colabeling of FL stem and progenitor populations, but not pulse at E8.5, the authors convincingly propose that ETPs are exclusively HSC−, and not YS−, derived. It is important to note the caveat, however, that while fate-mapping is a very powerful approach, many lineage-tracing models are both imperfect and overlapping. Deployment of pulse labeling within different developmental windows will and can suggest disparate origins depending on precisely when the label is induced and how the data are interpreted. There is still a need to identify more specific molecular markers of distinct embryonic progenitors so as to more precisely distinguish between the various waves of immune development during embryogenesis. Nonetheless, the work presented by Elsaid et al reflects a substantial contribution to understanding the mechanisms underlying developmental immune lrefayering and how layered immunity contributes to the generation of a functional immune system.
Conflict-of-interest disclosure: The authors declare no competing financial interests.