Abstract
Introduction: Chronic myelomonocytic leukaemia (CMML) is a clonal haematological neoplasm characterised by persistent monocytosis and myeloid dysplasia. Treatment options are few and hampered by incomplete understanding of its core biology. CMML is genetically homogeneous compared to most cancers, with >90% of patients displaying recurrent mutations in a small group of epigenetic regulator genes. Despite this, CMML exhibits substantial clinical heterogeneity, suggesting an important role for epigenetic dysregulation in CMML biology. However, the CMML epigenome remains little studied.
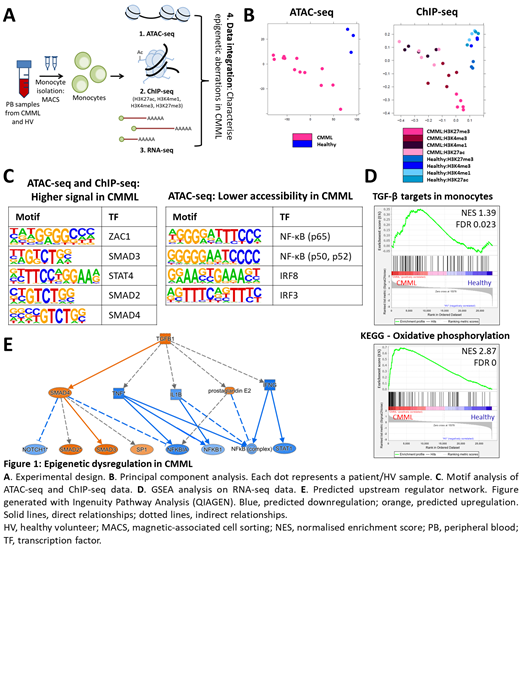
Methods: We performed multi-omic analyses on primary CD14 + monocytes from up to 13 CMML patients and 3 age-matched healthy controls, to identify regions of epigenetic dysregulation unique to CMML. Monocytes represent the defining downstream malignant cell population in CMML, contributing important disease features and supportive crosstalk with disease-initiating CMML stem cells; their targeting thus has therapeutic potential. We integrated RNA-seq, ATAC-seq and ChIP-seq for four histone marks, encompassing both activating and repressive marks (Fig 1A), to evaluate CMML monocytes at both the chromatin and transcriptome levels.
Results: All datasets clearly separated CMML from control monocytes by principal component analysis, whilst revealing substantial epigenetic heterogeneity between patients (Fig 1B). Most of the differentially accessible regions were distal to genes, suggestive of widespread enhancer dysregulation in CMML. ROSE analysis identified novel superenhancers specific to CMML monocytes, including several mapping to genes previously implicated in CMML biology (e.g. CXCL8).
Further analysis of differentially bound or accessible regions suggested consistent dysregulation of various pathways, including JAK/STAT, AKT and TREM1 signalling and the DNA damage response. Notably, there was strong epigenetic activation of the TGF-β pathway, with motifs for SMAD2, SMAD3, SMAD4 and FOXH1 consistently and strongly enriched across multiple datasets (Fig 1C, left). A signature of TGF-β target genes in monocytes, including many pro-survival genes, was also enriched in the matched RNA-seq data, suggesting a role for TGF-β activation in CMML monocytosis (Fig 1D, top). TGF-β signalling has been implicated in the monocyte-to-macrophage transition, but not previously as a driver of CMML biology.
Concurrently, there was strong epigenetic downregulation of the NF-κB pathway, evidenced by loss of chromatin accessibility at NF-κB binding elements in CMML monocytes (Fig 1C, right). This suggests a block in the inflammatory response of monocytes, expected to result in a tolerant phenotype. The block in NF-κB signalling was not directly evident in the RNA-seq data, likely reflecting absence of inflammatory stimuli at sampling. Extensive crosstalk between TGF-β and NF-κB signalling is recognised, implicating TGF-β activation in the observed repression of the NF-κB pathway in our data. A tolerant phenotype in monocytes has been previously linked to increased mitochondrial biogenesis. Indeed, RNA-seq highlighted higher expression of genes encoding components of the oxidative phosphorylation machinery in CMML monocytes (Fig 1D, bottom), including ATP5J, COX7A2 and NDUFB1.
Discussion: Combined transcriptomic and epigenomic analysis revealed profound dysregulation of the epigenetic landscape and of multiple signalling pathways in primary CMML monocytes. Whereas the ATAC-seq and ChIP-seq datasets aligned closely, significant discordance from the RNA-seq demonstrates the value of integrating multiple approaches for a complete picture of epigenetic dysregulation. Discordant changes identified at the chromatin but not transcriptomic level likely reflect poised potential. We describe TGF-β pathway activation in CMML for the first time, highlighting a potentially tractable therapeutic strategy. We propose a model whereby TGF-β activation directly represses NF-κB signalling potential in these cells, promoting a tolerant phenotype whilst conferring resistance to apoptosis (Fig 1E). This may be germane to the immune dysfunction (and propensity to autoimmunity) characteristic of CMML. Validation of lead candidate targets will be presented, highlighting novel therapeutic approaches for this disease of unmet clinical need.
Somervaille: Novartis: Consultancy, Honoraria. Wiseman: Bristol Myers Squibb: Consultancy; Astex: Research Funding; StemLine: Consultancy; Novartis: Consultancy; Takeda: Consultancy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal