Abstract
Introduction. Myeloproliferative neoplasms (MPN) are clonal disorders of hematopoietic stem cells that include polycythemia vera, essential thrombocythemia, and primary myelofibrosis. 10-20% of MPN pts transform to secondary acute myeloid leukemia (sAML), unresponsive to conventional therapy and associated with dismal outcome (Dunbar A, 2020). In addition to somatic driver mutations affecting JAK2, CALR or MPL, several additional variants are harbored by MPN pts inb chronic phase, and a restricted set of them were associated with risk of leukemic evolution (Vannucchi AM, Leukemia 2013; Tefferi A, Blood ASdv 2016) However, the molecular mechanisms underlying leukemic transformation remain largely unknown. Although bulk next generation sequencing (NGS) highlights the overall mutation landscape, it cannot distinguish which mutations occur in the same clone(s), nor elucidate the order of mutations or resolve clonal complexity. Conversely, single-cell sequencing (SCS) might allow to resolve clonal heterogeneity and reconstruct clonal phylogenies at each disease phase (Parenti, NPJ Prec Onc 2021)).
Aim: To delineate the clonal landscape of sAML, we performed single-cell mutational profiling in 10 pts with MPNs who progressed to sAML.
Methods. There were 2 set of samples/approaches: (i) 15 paired samples (chronic (CP)/blast phase (BP)) from 7 pts were analyzed using the Mission-Bio Tapestri platform and the Myeloid panel in order to target SNVs and indels across 45 myeloid genes with 312 amplicons. In one pt we also analyzed an intermediate phase corresponding to progression from PV to PPV-MF before BP development. (ii) 7 further paired CD34+ samples from 3 pts were analyzed using a 239-amplicon custom panel including 29 genes frequently mutated in myeloid neoplasms. SCS libraries were sequenced on Illumina Novaseq. Data were processed by using Mission Bio's Tapestri Pipeline and analyzed with Mission Bio's Tapestri Insights software package and R software. CNV analysis was performed by using an integrated pipeline for multiomics analysis (Mosaic, Mission Bio)
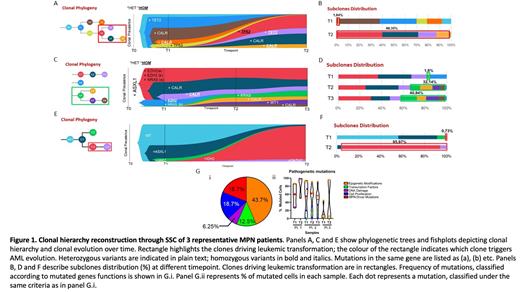
Results. (i) A total of 78,354 single cells were sequenced (average 5,223) using Tapestri Myeloid panel, with an average of 28,303 reads per cell and coverage of 97X. SCS was able to identify 17 low-frequency variants not detected in bulk analysis; however, it failed to discriminate homopolymeric regions including the ASXL1 G646Wfs*12. (ii) A total of 25,417 single cells were sequenced (average 3636) using a custom panel, with a coverage of 186X and an average Allele Dropout Rate of 8.6%. This panel was able to identify ASXL1 G646Wfs*12 variant. Overall, we found a significant correlation of variant allele frequency (VAF) measured by bulk and SCs approach (R =0.84, p<.0001). Epigenetic variants (i.e. ASXL1, TET2, EZH2) account for around half of the mutations and affect a large fraction of CP cells (3 representative samples in Fig.1). In 8/10 pts, leukemic clone emerged from a driver mutation-positive cells (JAK2V617F n=4; CALR Type1 n=4). In all pts we are able to identify at least 3 mutated clones and in 7 pts the dynamics of the clones allowed to identify the one(s) responsible for evolution to sAML. In 7/10 pts, the leukemic clones were already detectable at low frequency (<2%) at CP and became dominant in BP; these low-frequency clones were missed by bulk sequencing. Furthermore, SCS revealed acquisition of 3 mutually exclusive mutations in RAS pathway in one pt: two NRAS mutations and a KRAS mutation. Copy number variation (CNV) could be assessed in 4 pts. Of note, during progression to sAML, we found single cells with amplification of ETV6 (>20 copies in 2 pts), NRAS (8 copies in 2pts) and BRAF (8 copies in 2pts). Other subclonal ploidy abnormalities were also observed in RUNX1, EZH2, U2AF1 and ZRSR2 (5-18 copies).
Conclusions. Together, these data suggest that MPN present a complex clonal combination evolving over time. SCS allows to resolve this milieu, that is largely missed by conventional bulk NGS, in particularly SCS identifies rare leukemia-driving clones that were already present in chronic phase and describes their dynamics during leukemic progression. Leukemic transformation after MPN is a highly heterogeneous process with mutations and CNVs acquired in different genes and different clones. Overall, our findings provide further insights into the pathogenesis of AML transformation of MPN.
Supported by AIRC, Mynerva project no.21267
Vannucchi: BMS: Honoraria, Membership on an entity's Board of Directors or advisory committees; Incyte: Honoraria, Membership on an entity's Board of Directors or advisory committees; AbbVie: Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal