Abstract
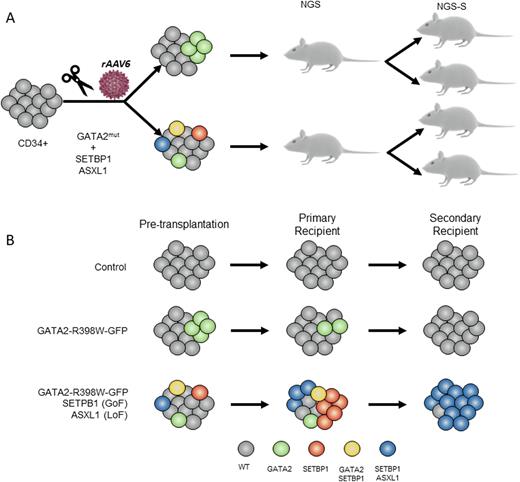
GATA2 deficiency has been identified as a common hereditary cause of myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML) in children. Penetrance and expressivity within affected families is often variable, suggesting that cooperating factors are required to trigger the disease. Somatic mutations in MDS driver genes (i.e. SETBP1, ASXL1) have been identified by us and others in GATA2-MDS. Despite recent studies identifying germline genetics and acquired mutations in GATA2 deficiency, the understanding of the molecular mechanism that triggers the leukemic progression remains still unknown. Particularly, it is still unanswered the following biological questions: 1) whether GATA2 germline mutation itself is sufficient to trigger MDS/AML 2) whether SETBP1 and ASXL1 mutations induce the malignant transformation. Addressing these questions has been challenging due to the absence of trustworthy human disease models. Here, we used CRISPR/Cas9 genome engineering of primary Cord Blood (CB) CD34+ cells followed by serial transplantation into immunodeficient mice to study the genetic malignant effect of GATA2 (loss-of-function R398W mutation), SETBP1 (gain-of-function D868N) and ASXL1 (loss-of-function G646X) mutations. Specifically, CRISPR/Cas9 in combination with recombinant adeno-associated virus serotype 6 (rAAV6) were used to knock-in a modified GATA2 exon 6 (exon6-T2A-GFP) carrying the R398W mutation in CB-CD34+ cells alone (referred to as GATA2) or in combination with mutations in SETBP1 and ASXL1 (TRIPLE). Mock-treated CD34+ cells were nucleofected without RNP, rAAV6 or ssODN (Control). Four days after the nucleofection, ~11% of cells were GFP positive in GATA2 condition (n=3) and ~7% in TRIPLE condition (n=3), which directly correlates with the percentage of GATA2-R398W mutant cells. Next, manipulated CD34+ cells (Control, GATA2 and TRIPLE) were transplanted into sublethally irradiated NSG mice (n=24). Sixteen weeks after transplantation, 18/24 mice showed more than 10% of human cell engraftment (hCD45) in the bone marrow (BM). Moreover, a multilineage reconstitution (hCD34 for HPCS, hCD33 for myeloid cells and hCD19 for lymphoid cells) was observed in all the conditions. In order to dissect the clonal architecture of concurrent mutations, we performed single-cell DNA-sequencing for GATA2, SETBP1 and ASXL1 in hundreds of human CD45+ engrafted cells derived from colony forming units (CFU). Fifty of a total of 118 colonies analyzed (43%) had targeted editing of at least one gene (GATA2, SETBP1 or ASXL1). Interestingly, in TRIPLE condition 13/51 (24%) colonies, more than one gene was edited. To study the long-term clonal evolution, we performed secondary transplants.Nine out of 15 mice reached the end point of 24 weeks.
We noted that the predominant expanded clones were carrying either SETBP1 or SETBP1+ASXL1 mutations, while clones with only GATA2 mutation were lost in both GATA2 and TRIPLE conditions. To study the impact of the mutations at transcriptomic level, single cell RNA-seq from primary and secondary hCD45+/CD19- engrafted cells was performed (the analysis is underway). In summary, we developed a new in vivo human model of GATA2 deficiency and GATA2-MDS-specific mutations by CRISPR/Cas9 targeting of CD34+ cells. Our findings strongly suggest that GATA2 R398W mutation is not sufficient to increase the fitness of the mutant cells, suggesting that a co-operation of genetic, epigenetic and stressor factors is necessary to trigger the clonal expansion.
Disclosures
Bödör:Takeda: Speakers Bureau; Epizyme: Speakers Bureau; Amgen: Speakers Bureau; Astra-Zeneca: Speakers Bureau; Astellas Pharma: Speakers Bureau; Janssen: Speakers Bureau; Novartis: Speakers Bureau; Abbvie: Research Funding; Roche: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal