Key Points
Baseline 18F-FDG–PET radiomics features can select patients at high risk more accurately than the IPI risk score.
The clinical PET model that was developed in the HOVON-84 data set remained predictive of the outcome in 6 independent studies.
Abstract
The objective of this study is to externally validate the clinical positron emission tomography (PET) model developed in the HOVON-84 trial and to compare the model performance of our clinical PET model using the international prognostic index (IPI). In total, 1195 patients with diffuse large B-cell lymphoma (DLBCL) were included in the study. Data of 887 patients from 6 studies were used as external validation data sets. The primary outcomes were 2-year progression-free survival (PFS) and 2-year time to progression (TTP). The metabolic tumor volume (MTV), maximum distance between the largest lesion and another lesion (Dmaxbulk), and peak standardized uptake value (SUVpeak) were extracted. The predictive values of the IPI and clinical PET model (MTV, Dmaxbulk, SUVpeak, performance status, and age) were tested. Model performance was assessed using the area under the curve (AUC), and diagnostic performance, using the positive predictive value (PPV). The IPI yielded an AUC of 0.62. The clinical PET model yielded a significantly higher AUC of 0.71 (P < .001). Patients with high-risk IPI had a 2-year PFS of 61.4% vs 51.9% for those with high-risk clinical PET, with an increase in PPV from 35.5% to 49.1%, respectively. A total of 66.4% of patients with high-risk IPI were free from progression or relapse vs 55.5% of patients with high-risk clinical PET scores, with an increased PPV from 33.7% to 44.6%, respectively. The clinical PET model remained predictive of outcome in 6 independent first-line DLBCL studies, and had higher model performance than the currently used IPI in all studies.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of aggressive non-Hodgkin lymphoma in adults with large variations in outcomes. Approximately 20% to 50% of patients with DLBCL are refractory to standard chemo-immunotherapy or relapse after achieving complete response.1 With more available innovative treatment options (such as chimeric antigen T-cell and bispecific monoclonal therapy), better selection of patients at high risk is highly relevant to potentially offer these patients a timely switch to these new treatment options.
Thirty years after its development, the international prognostic index (IPI)2 is still the most widely used prognostic index for DLBCL. The addition of rituximab has significantly increased the cure rate.3 The ability to identify patients at high risk with a long-term survival of <50% using the IPI, revised IPI, and National Comprehensive Cancer Network IPI is limited.4,5 Therefore, more accurate prognostic markers are essential to identify patients at high risk of progression or relapse. In recent years, several studies have explored the potential of the baseline metabolic tumor volume (MTV) extracted from 18F-fluorodeoxyglucose positron emission tomography–computed tomography (18F-FDG–PET/CT) scans to predict the DLBCL outcome. The results consistently showed that MTV is inversely related to overall survival and progression-free survival (PFS).6-11 Recently, a new international prognostic index (IMPI) incorporating MTV, age, and Ann Arbor stage was developed, thereby allowing improved individual outcome prediction.12
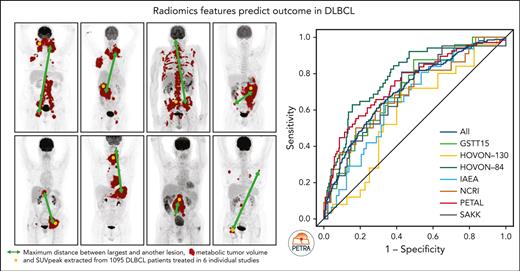
MTV reflects the 18F-FDG–avid tumor burden but does not include phenotypical aspects such as the spatial distribution, heterogeneity, and shape of lesions. Recently developed quantitative 18F-18F-FDG–PET/CT features, also referred to as radiomics, reveal the biological characteristics of the disease and could help to improve outcome prediction. Adding 18F-FDG–PET radiomics features to the currently used predictors may improve the identification of patients with poor prognosis. Features quantifying dissemination, in particular, have shown high predictive value independent from MTV in DLBCL.11,13 Therefore, we previously developed a prediction model that incorporated MTV, the peak of the standardized uptake value (SUVpeak), the maximum distance between the largest lesion and any other lesion (Dmaxbulk), World Health Organization (WHO) performance status, and age using data of the HOVON-84 trial.11 The advantage of this model over other models using dichotomous cutoffs is that it allows for individual patient risk prediction and is less sensitive to data-driven cutoffs.
The objective of this study is to externally validate the clinical positron emission tomography (PET) model developed in the HOVON-84 trial11 using 887 patients from the PETRA database and to compare the model performance of our clinical PET model with the currently used IPI.
Methods
Study population
Adult patients with de novo DLBCL (n = 1466) with a baseline 18F-FDG–PET scan and 2-year follow-up data were included. Clinical data and [18F]FDG-PET scans were collated and harmonized by the PETRA consortium.14 Patients were originally included in 7 individual studies: GSTT15,7 HOVON-84,15 HOVON-130,16 IAEA,17 NCRI,18 PETAL,19 and SAKK 38/0720 (hereafter referred to as SAKK). Individual trials were approved by the institutional review board and all patients provided written informed consent. The use of all data within the PETRA imaging database was approved by the institutional review board of VU University Medical Center (JR/20140414).
18F-FDG–PET/CT analysis
Scans did not pass quality control if (1) whole body 18F-FDG–PET/CT scans were incomplete, (2) essential Digital Imaging and Communications in Medicine (DICOM) information was missing, (3) no FDG-avid lesions were present, and (4) plasma glucose levels and hepatic SUVmean were outside the suggested ranges of the European Association of Nuclear Medicine.21 Scans were included when the hepatic SUVmean was outside the suggested ranges, but the total image activity was between 50% and 80% of the total injected activity.
Quantitative analysis of all 18F-FDG–PET scans that passed quality control was performed using the ACCURATE tool.22 Lesions were delineated at baseline using a fully automated preselection defined by SUV ≥4.0, and a volume threshold ≥3 mL.23 Previous studies showed that an SUV threshold of 4.0 and a volume threshold of ≥3 mL resulted in the highest success rate and interobserver variability.23,24 Physiological uptake was deleted, and lymphoma lesions <3 mL were added with single mouse clicks. The physiological uptake (eg, bladder and kidneys) adjacent to the tumor regions was removed manually. All scans were reviewed by a nuclear medicine physician who was blinded to the outcome. Delineations were performed by a nuclear medicine physician (GSTT15 and IAEA) or under the supervision of a nuclear medicine physician by trained researchers (with >5 years of experience; HOVON-84, HOVON-130, PETAL, NCRI, and SAKK). We assessed the concordance of MTV between a nuclear medicine physician and a trained researcher for the SAKK study, and observed a correlation of 0.99.12 To further harmonize quantitative 18F-FDG–PET analysis between studies, all segmentations were visually checked for missed lesions or missed physiological uptake by a trained researcher before calculating the radiomics features. Based on these delineations, the MTV, SUVpeak,25 and Dmaxbulk were extracted for all patients. During model development using the HOVON-84 trial, we choose SUVpeak instead of SUVmax because the SUVpeak is relatively less sensitive to noise.26 All image-processing and feature calculations were performed using RaCaT software,27 which is in compliance with the imaging biomarker standardization initiative criteria.28
Statistical analysis
Prediction models
Multivariable logistic regression with backward feature selection was used to predict the risk of progression, relapse, or death after 2 years (2-year PFS) and the risk of progression or relapse after 2 years (2-year time to progression [TTP]). Follow-up started at the time of baseline [18F]FDG–PET/CT scan. Patients who died within 2 years without signs of progression or relapse were excluded from the TTP prediction model.
We tested the predictive value of the following models:
IPI: the IPI risk score using low, low-intermediate, high-intermediate and high-risk groups.2
Clinical PET model as developed in the HOVON-84 trial: the natural logarithms of MTV and SUVpeak, the maximum distance between the largest lesion and any other lesion (Dmaxbulk), WHO performance status, and age.11
For the clinical PET model, the sum of individual predictors, weighted based on regression coefficients, together with the intercept of the model, were used to derive the predicted probability of an event for each patient. The model performance was assessed using the area under the curve (AUC) of the receiver operating characteristic curve. Differences between the model performances of prediction models, expressed as AUC, were assessed using the two-sided DeLong test.29
Updating the model
Ideally, a prediction model provides valid predictions of the outcome for individual patients in a setting other than that in which the model was developed. Recalibration methods for reestimating the coefficients of a model are attractive because of their stability. The validity of the model predictions can be assessed by comparing the observed outcomes and predictions when empirical data from this external setting are available,30 which is the case now that we have 887 patients available from 6 external studies. We updated the model using all available data within the PETRA using logistic calibration. The intercept was updated to make the average predicted probability equal to the observed overall event rate (so-called calibration-in-the-large), and individual coefficients were reestimated.30 Detection of calibration-in-the-large problems avoids miscalibration of the model and, consequently, wrong decision making.30
Sensitivity analysis
We assessed model performances among patients exclusively treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP). Secondly, we investigated the added value of the cell of origin (COO) to our clinical PET prediction model in a subset of patients with available COO information.
Furthermore, to compare the model performance of our clinical PET model with that of the IMPI model12 and a model that combined MTV and WHO performance status (MTV/ECOG),31 we applied Cox regression models with a 2-year PFS as the outcome and assessed model performance, using the C-index and the Akaike information criteria.
Diagnostic performance
To calculate the diagnostic performance of the models, high- and low-risk groups were defined. For the IPI prediction model, patients with 4 or 5 adverse factors were considered as high risk. For the clinical PET model, patients with the highest predicted probabilities were used to define the high-risk group. To allow comparison of the high-risk groups of the IPI and clinical PET models, the high-risk patient group for the clinical PET model was of equal size to the high-risk IPI group. The diagnostic performance of the prediction models was assessed using sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). For the Cox regression models, high-risk groups for the IMPI and clinical PET models were of equal size as the high-risk IPI group and the MTV/ECOG group with 2 risk points. Survival curves were obtained with Kaplan-Meier analyses, using the probabilities of the Cox regression models to create risk groups.
Statistical analysis was performed using R (version 4.2.1). P < .05 was considered statistically significant.
Results
Patient characteristics
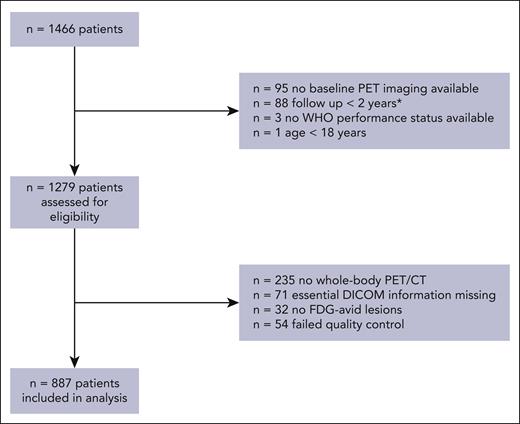
A total 1466 eligible patients with de novo DLBCL from studies other than the HOVON-84 study were available in the PETRA database, of whom 887 were included in this analysis (Figure 1). Patients with no baseline 18F-FDG–PET imaging available (n = 95), who were lost to follow-up within 2 years and did not show any signs of progression (n = 88), aged <18 years (n = 1), and with missing WHO performance status (n = 3) were ineligible for this study. 18F-FDG–PET quality control led to the exclusion of patients with incomplete 18F-FDG–PET/CT scans (n = 235), missing essential DICOM information (n = 71), no 18F-FDG–avid lesions (n = 32), and scans outside the quality control range (n = 54). For the Cox regression models, patients who had a follow-up shorter than 2 years and an 18F-FDG–PET/CT scan that was within our quality control were included (n = 58).
CONSORT diagram of included patients for external validation. ∗Patients who were not included in the logistic regression model but were included in the Cox regression model.
CONSORT diagram of included patients for external validation. ∗Patients who were not included in the logistic regression model but were included in the Cox regression model.
Together with 308 patients from the HOVON-84 study, a total of 1195 patients were included in this analysis. Descriptive statistics of the baseline characteristics of all included patients stratified per the study are presented in Table 1. Two hundred and forty-one patients developed progression or relapse within 2 years after baseline 18F-FDG–PET/CT, and 50 patients died within 2 years after baseline 18F-FDG–PET/CT. The median baseline MTV of all patients was 324.4 mL (interquartile range [IQR], 81.7-828.8), with a median SUVpeak of 17.6 (IQR, 12.1-24.4) and a median Dmaxbulk of 22.2 cm (4.8-41.2; supplemental Table 1, available on the Blood website).
Characteristics of included patients
| . | Total (n = 1195) . | GSTT157 (n = 97) . | HOVON-13016 (n = 65) . | HOVON-8415 (n = 308) . | IAEA17 (n = 104) . | NCRI18 (n = 133) . | PETAL19 (n = 368) . | SAKK20 (n = 120) . |
|---|---|---|---|---|---|---|---|---|
| Age (median, IQR) | 62 (51-70) | 61 (49-70) | 63 (54-72) | 65 (56-72) | 57 (43-65) | 61 (49-68) | 61 (51-70) | 59 (49-68) |
| >60 y | 547 (46) | 47 (48) | 30 (46) | 100 (32) | 63 (61) | 63 (47) | 179 (49) | 65 (54) |
| ≤60 y | 648 (54) | 50 (52) | 35 (54) | 208 (68) | 41 (39) | 70 (53) | 189 (51) | 55 (46) |
| Ann Arbor stage | ||||||||
| I | 108 (9) | 9 (9) | 0 | 0 | 11 (11) | 8 (6) | 66 (18) | 14 (12) |
| II | 284 (24) | 20 (21) | 7 (11) | 55 (18) | 25 (24) | 51 (38) | 80 (22) | 42 (35) |
| III | 269 (23) | 11 (11) | 8 (12) | 70 (23) | 23 (22) | 35 (26) | 75 (20) | 26 (22) |
| IV | 534 (45) | 57 (59) | 50 (77) | 183 (59) | 45 (43) | 39 (29) | 147 (40) | 38 (32) |
| WHO performance status | ||||||||
| 0 | 590 (49) | 32 (33) | 38 (58) | 175 (57) | 36 (35) | 75 (56) | 166 (45) | 68 (57) |
| 1 | 449 (38) | 35 (36) | 23 (35) | 94 (31) | 44 (42) | 44 (33) | 165 (45) | 44 (37) |
| 2 | 124 (10) | 18 (19) | 3 (5) | 39 (13) | 15 (14) | 14 (11) | 27 (7) | 8 (7) |
| 3 | 30 (3) | 12 (12) | 1 (2) | 0 | 7 (7) | 0 | 10 (3) | 0 |
| 4 | 2 | 0 | 0 | 0 | 2 (2) | 0 | 0 | 0 |
| LDH | ||||||||
| ≤ Normal | 478 (40) | 35 (36) | 16 (25) | 100 (32) | 54 (52) | 51 (38) | 154 (42) | 62 (52) |
| Normal | 713 (60) | 62 (64) | 45 (69) | 208 (68) | 50 (48) | 82 (62) | 214 (58) | 58 (48) |
| Missing | 4 | 4 (6) | ||||||
| Extranodal involvement | ||||||||
| ≥1 | 773 (65) | 47 (48) | 30 (46) | 182 (59) | 67 (64) | 106 (80) | 249 (68) | 92 (77) |
| <1 | 422 (35) | 50 (52) | 35 (54) | 126 (41) | 37 (36) | 27 (20) | 119 (32) | 28 (23) |
| IPI low | 368 (31) | 26 (27) | 9 (14) | 51 (17) | 44 (42) | 52 (39) | 125 (34) | 61 (51) |
| Low-intermediate | 264 (22) | 10 (10) | 14 (22) | 75 (24) | 16 (15) | 28 (31) | 97 (26) | 24 (20) |
| High-intermediate | 331 (28) | 30 (31) | 29 (45) | 106 (34) | 22 (21) | 35 (26) | 89 (24) | 20 (17) |
| High | 232 (19) | 31 (32) | 13 (20) | 76 (25) | 22 (21) | 18 (14) | 57 (15) | 15 (13) |
| . | Total (n = 1195) . | GSTT157 (n = 97) . | HOVON-13016 (n = 65) . | HOVON-8415 (n = 308) . | IAEA17 (n = 104) . | NCRI18 (n = 133) . | PETAL19 (n = 368) . | SAKK20 (n = 120) . |
|---|---|---|---|---|---|---|---|---|
| Age (median, IQR) | 62 (51-70) | 61 (49-70) | 63 (54-72) | 65 (56-72) | 57 (43-65) | 61 (49-68) | 61 (51-70) | 59 (49-68) |
| >60 y | 547 (46) | 47 (48) | 30 (46) | 100 (32) | 63 (61) | 63 (47) | 179 (49) | 65 (54) |
| ≤60 y | 648 (54) | 50 (52) | 35 (54) | 208 (68) | 41 (39) | 70 (53) | 189 (51) | 55 (46) |
| Ann Arbor stage | ||||||||
| I | 108 (9) | 9 (9) | 0 | 0 | 11 (11) | 8 (6) | 66 (18) | 14 (12) |
| II | 284 (24) | 20 (21) | 7 (11) | 55 (18) | 25 (24) | 51 (38) | 80 (22) | 42 (35) |
| III | 269 (23) | 11 (11) | 8 (12) | 70 (23) | 23 (22) | 35 (26) | 75 (20) | 26 (22) |
| IV | 534 (45) | 57 (59) | 50 (77) | 183 (59) | 45 (43) | 39 (29) | 147 (40) | 38 (32) |
| WHO performance status | ||||||||
| 0 | 590 (49) | 32 (33) | 38 (58) | 175 (57) | 36 (35) | 75 (56) | 166 (45) | 68 (57) |
| 1 | 449 (38) | 35 (36) | 23 (35) | 94 (31) | 44 (42) | 44 (33) | 165 (45) | 44 (37) |
| 2 | 124 (10) | 18 (19) | 3 (5) | 39 (13) | 15 (14) | 14 (11) | 27 (7) | 8 (7) |
| 3 | 30 (3) | 12 (12) | 1 (2) | 0 | 7 (7) | 0 | 10 (3) | 0 |
| 4 | 2 | 0 | 0 | 0 | 2 (2) | 0 | 0 | 0 |
| LDH | ||||||||
| ≤ Normal | 478 (40) | 35 (36) | 16 (25) | 100 (32) | 54 (52) | 51 (38) | 154 (42) | 62 (52) |
| Normal | 713 (60) | 62 (64) | 45 (69) | 208 (68) | 50 (48) | 82 (62) | 214 (58) | 58 (48) |
| Missing | 4 | 4 (6) | ||||||
| Extranodal involvement | ||||||||
| ≥1 | 773 (65) | 47 (48) | 30 (46) | 182 (59) | 67 (64) | 106 (80) | 249 (68) | 92 (77) |
| <1 | 422 (35) | 50 (52) | 35 (54) | 126 (41) | 37 (36) | 27 (20) | 119 (32) | 28 (23) |
| IPI low | 368 (31) | 26 (27) | 9 (14) | 51 (17) | 44 (42) | 52 (39) | 125 (34) | 61 (51) |
| Low-intermediate | 264 (22) | 10 (10) | 14 (22) | 75 (24) | 16 (15) | 28 (31) | 97 (26) | 24 (20) |
| High-intermediate | 331 (28) | 30 (31) | 29 (45) | 106 (34) | 22 (21) | 35 (26) | 89 (24) | 20 (17) |
| High | 232 (19) | 31 (32) | 13 (20) | 76 (25) | 22 (21) | 18 (14) | 57 (15) | 15 (13) |
LDH, lactate dehydrogenase.
Prediction model
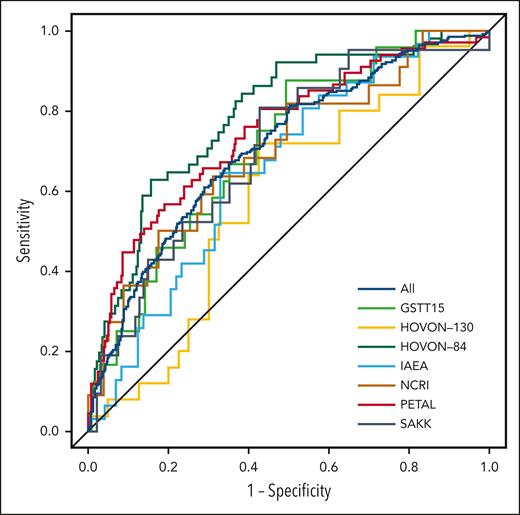
Using a 2-year PFS as the outcome, the AUC of the HOVON-84 trial was 0.67 for the IPI model and 0.75 for the clinical PET model.11 The IPI model yielded an AUC of 0.62 using all patients (Table 2; Figure 2). Within individual studies, the AUC of the IPI model ranged from 0.51 for the SAKK study to 0.65 for the PETAL study. The clinical PET model yielded an AUC of 0.71, which was significantly higher than that of the IPI model (P < .001). The AUC of the clinical PET model ranged between 0.59 for the HOVON-130 study to 0.75 for the PETAL study. For all individual studies, the AUC of the clinical PET model was higher than that of the IPI model, especially for the IAEA and SAKK studies.
AUCs of the IPI prediction model and clinical PET prediction models for all individual studies and all patients using 2-year PFS and 2-year TTP as the outcomes
| Study name . | 2-y PFS . | 2-y TTP . | ||
|---|---|---|---|---|
| IPI . | Clinical PET . | IPI . | Clinical PET . | |
| HOVON-84 (test) | 0.67 | 0.75 | 0.69 | 0.79 |
| All patients | 0.62 | 0.71 | 0.62 | 0.71 |
| GSTT15 | 0.63 | 0.72 | 0.62 | 0.71 |
| HOVON-130 | 0.53 | 0.59 | 0.53 | 0.60 |
| IAEA | 0.56 | 0.65 | 0.56 | 0.66 |
| NCRI | 0.56 | 0.71 | 0.59 | 0.70 |
| PETAL | 0.65 | 0.75 | 0.62 | 0.75 |
| SAKK | 0.51 | 0.71 | 0.51 | 0.70 |
| Study name . | 2-y PFS . | 2-y TTP . | ||
|---|---|---|---|---|
| IPI . | Clinical PET . | IPI . | Clinical PET . | |
| HOVON-84 (test) | 0.67 | 0.75 | 0.69 | 0.79 |
| All patients | 0.62 | 0.71 | 0.62 | 0.71 |
| GSTT15 | 0.63 | 0.72 | 0.62 | 0.71 |
| HOVON-130 | 0.53 | 0.59 | 0.53 | 0.60 |
| IAEA | 0.56 | 0.65 | 0.56 | 0.66 |
| NCRI | 0.56 | 0.71 | 0.59 | 0.70 |
| PETAL | 0.65 | 0.75 | 0.62 | 0.75 |
| SAKK | 0.51 | 0.71 | 0.51 | 0.70 |
Receiver operating characteristic curves for 2-year PFS for all included patients and separate studies.
Receiver operating characteristic curves for 2-year PFS for all included patients and separate studies.
Comparable results were obtained using a 2-year TTP as the outcome. The AUC of the HOVON-84 trial for IPI was 0.69, vs 0.79 for the clinical PET model. The IPI model yielded an AUC of 0.62, and the clinical PET model yielded an AUC of 0.71, when using all patients (P < .001). Again, for all individual studies, the AUCs of the clinical PET models were consistently higher than the AUCs of the IPI model.
Diagnostic performance
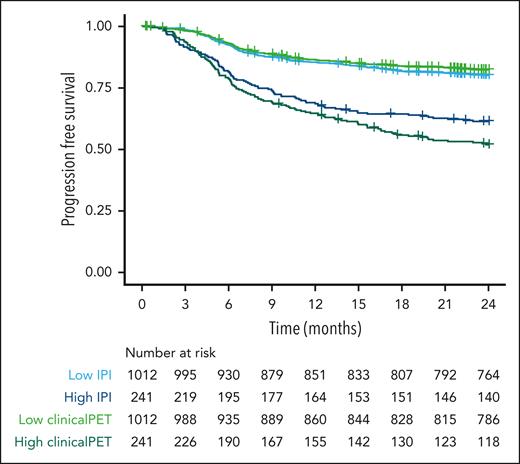
Patients at high risk according to the IPI model had a 2-year PFS probability of 61.4% (95% confidence interval [CI], 55.5-67.9; Figure 3). Patients at high risk according to the clinical PET model had a probability for 2-year PFS of 51.9% (95% CI, 45.9-58.7). The sensitivity, specificity, PPV, and NPV were higher for the clinical PET model than for the IPI model (Table 3). Specificity and NPV showed a small increase, but sensitivity increased from 29.5% to 39.0%, and PPV increased from 35.5% in the IPI model to 49.1% in the clinical PET model.
Survival curves of patients at high and low risk, as identified with IPI and clinical PET models, using 2-year PFS as the outcome.
Survival curves of patients at high and low risk, as identified with IPI and clinical PET models, using 2-year PFS as the outcome.
Diagnostic performance of the IPI and clinical PET models
| . | . | Sensitivity (95% CI) . | Specificity (95% CI) . | PPV (95% CI) . | NPV (95% CI) . |
|---|---|---|---|---|---|
| PFS | IPI | 27.90 (22.69-33.59) | 84.51 (81.99-86.81) | 35.48 (30.13-41.23) | 79.34 (78.02-80.59) |
| Clinical PET | 39.18 (33.53-45.04) | 86.95 (84.57-89.08) | 49.14 (43.65-54.65) | 81.62 (80.14-83.01) | |
| TTP | IPI | 29.46 (23.78-35.65) | 84.51 (81.99-86.81) | 33.65 (28.36-39.38) | 81.80 (80.48-83.05) |
| Clinical PET | 39.00 (32.81-45.47) | 87.06 (84.69-89.18) | 44.55 (38.93-50.31) | 84.26 (82.83-85.59) |
| . | . | Sensitivity (95% CI) . | Specificity (95% CI) . | PPV (95% CI) . | NPV (95% CI) . |
|---|---|---|---|---|---|
| PFS | IPI | 27.90 (22.69-33.59) | 84.51 (81.99-86.81) | 35.48 (30.13-41.23) | 79.34 (78.02-80.59) |
| Clinical PET | 39.18 (33.53-45.04) | 86.95 (84.57-89.08) | 49.14 (43.65-54.65) | 81.62 (80.14-83.01) | |
| TTP | IPI | 29.46 (23.78-35.65) | 84.51 (81.99-86.81) | 33.65 (28.36-39.38) | 81.80 (80.48-83.05) |
| Clinical PET | 39.00 (32.81-45.47) | 87.06 (84.69-89.18) | 44.55 (38.93-50.31) | 84.26 (82.83-85.59) |
For 2-year TTP as the outcome, patients with high-risk IPI scores had a survival rate of 66.4% (95% CI, 60.3-73.0). Patients with high-risk clinical PET scores had a survival rate of 55.5% (95% CI, 49.1-62.6). Again, sensitivity, specificity, PPV, and NPV were higher for the clinical PET than for the IPI model. The PPV increased from 33.7% to 44.6% in the clinical PET model compared with that in the IPI model.
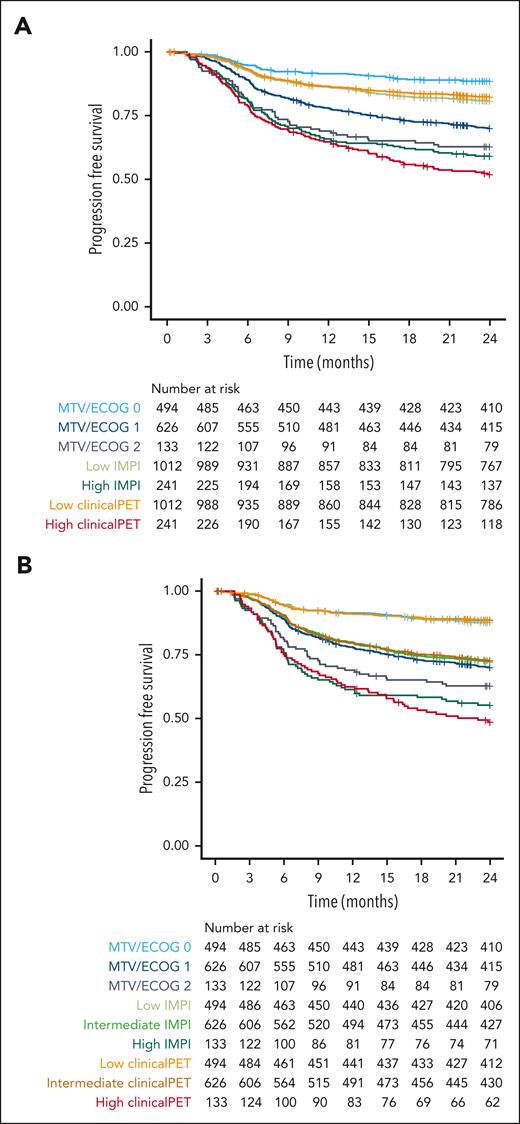
Patients with 2 risk points in the MTV/ECOG model had a 2-year PFS of 62.8% (95% CI, 55.0-71.6; Figure 4), whereas patients at high risk according to the IMPI scores had a 2-year PFS of 59.1% (95% CI, 53.2-65.7). Patients at high risk according to the clinical PET model had the lowest survival rate, with a 2-year PFS of 51.9% (95% CI, 45.9-58.7). When using the same group sizes for the high-risk group as those of the patients with 2 risk points in the MTV/ECOG model, the 2-year PFS rates of the patients at high risk according to the IMPI scores were 55.2% (95% CI, 47.4-64.4) and 48.6% (95% CI, 40.8-57.9) using the clinical PET model, showing a clear superiority of both the IMPI and clinical PET model, with the best selection of patients at high risk by the clinical PET model, which is in line with the C-index and AIC values of the models.
Survival curves of patients at high and low risk as identified with MTV/ECOG, IMPI, and clinical PET models using 2-year progression-free survival as outcome. (A) Risk groups of the MTV/ECOG as defined in the original publication32 and high-risk groups of the IMPI and clinical PET models are of equal size as the high-risk IPI group. (B) Risk groups for all models are of equal size as the MTV/ECOG groups.
Survival curves of patients at high and low risk as identified with MTV/ECOG, IMPI, and clinical PET models using 2-year progression-free survival as outcome. (A) Risk groups of the MTV/ECOG as defined in the original publication32 and high-risk groups of the IMPI and clinical PET models are of equal size as the high-risk IPI group. (B) Risk groups for all models are of equal size as the MTV/ECOG groups.
Updating the model
After updating the model, its model performance (supplemental Table 2) and diagnostic performance (supplemental Table 3) were comparable with those of the original HOVON-84 model. For the GSTT, PETAL, and NCRI studies, the model performance slightly improved after calibration, whereas it decreased for the HOVON-130, IAEA, and SAKK studies. The diagnostic performance was slightly higher after model recalibration.
Sensitivity analysis
Similar results were obtained when only patients treated with R-CHOP were included (n = 1157 patients). The performance of the clinical PET model increased for the GSTT15, IAEA, and PETAL studies (supplemental Table 2). For both 2-year PFS and 2-year TTP, the AUC of IPI was 0.62, and that our clinical PET model was 0.71. A total of 493 patients had COO information available. In this subset, the COO was not a significant predictor of outcome after backward feature selection.
Furthermore, Cox regression modeling showed that model performance was highest for the clinical PET model (C-index, 0.69) and lowest for the MTV/ECOG model (C-index, 0.63); IMPI had a C-index of 0.66. Similar results were observed for the AIC (supplemental Table 4).
Discussion
Our study shows that the clinical PET model that was developed in the HOVON-84 trial remained predictive of outcome in 6 independent studies and had better model performance than the currently used IPI in all studies. Baseline 18F-FDG–PET clinical PET features were superior to IPI in identifying patients with high-risk DLBCL, with a relatively better model performance and higher PPV.
Several other studies have evaluated the predictive value of baseline radiomics features in DLBCL.11,32-38 Because of the different (numbers of) features that were extracted, it is hard to compare these studies directly. In general, all studies confirm that radiomics features are predictive of outcome. Moreover, previous studies showed that dissemination is a predictor of outcome independent of MTV.13,32 A recent study compared the 3 IPI variants in 2124 patients; according to the original IPI, patients had a 2-year PFS of almost 60%,5 which is comparable to the IPI performance in our study.
Cottereau et al32 published a risk stratification model that included the maximum distance between 2 lesions normalized for the body surface area (SDmax) and MTV in 301 patients. They showed that patients with both high MTV and SDmax had significantly lower survival rates, with a 2-year PFS of ∼50%. These results are comparable with our results, given that we reported a 2-year PFS of 51.9% in the high-risk group. Both high-risk groups included ∼20% of the patients. However, it should be noted that they applied a different segmentation method to delineate lesions, which could probably explain the lower median MTV (253 mL vs 324.4 mL) and hampers direct comparison of their model to ours, because multiple studies have shown large differences in extracted MTVs using the SUV4.0 or 41% max segmentation methods.6,24,39 Previous analysis in the HOVON-84 study showed that correction of Dmaxbulk for height did not influence our model performance.11 Moreover, the advantage of our clinical PET model is that it allows individual patient risk prediction because MTV and Dmaxbulk are included as continuous variables. Therefore, it is less influenced by data-driven optimal cutoffs. A dichotomous cutoff results in different survival estimates for MTV and SDmax values that are close to the cutoffs, whereas the actual survival is similar and more accurately predicted with our clinical PET model.
Kostakoglu et al40 recently published a radiomics prediction model based on 1263 patients from the GOYA trial. Patient characteristics were comparable, although their study included patients with slightly more advanced-stage diseases (84% vs 68%, respectively), and our study included more patients with high-risk IPI (15% vs 19%, respectively). Although their model performance was lower (AUC 0.64), the patients at high risk (33% of the total population), which their random forest prediction model identified, had a 2-year PFS of ∼50%. In this study, 42 radiomics features were used. In addition to the MTV, 7 textural features were included in the final random forest model. Textural features are sensitive to different acquisition, reconstruction, and segmentation methods,39,41,42 leading to limited reproducibility in multicenter, multivendor studies, which was the case for 5 out of the 7 textural features included in their prediction model.42 Moreover, interpretation of these textural features is complex. Contrary to textural radiomics features, dissemination features are easy to interpret because they quantitatively reflect what can be visualized using 18F-FDG–PET/CT scans. They are also relatively simple to calculate and are relatively insensitive to scan protocol differences.
The recently published IMPI included Ann Arbor stage, age, and MTV.12 In our clinical PET model, Ann Arbor stage is replaced by Dmaxbulk and WHO performance status. Both IMPI and clinical PET models allow individual risk prediction. Looking at the 2-year PFS rates, the clinical PET model outperformed both IMPI and MTV/ECOG prediction models.
None of the previously described prognostic models reported the PPV, NPV, sensitivity, and specificity; therefore, we cannot compare the diagnostic measures of these radiomics models with those of our clinical PET model. The high-risk groups in all the mentioned prediction models and our clinical PET model had a survival rate of ∼50%, indicating that none of the indices identified a truly high-risk group. There is an unmet need to identify patients with high-risk DLBCL shortly after diagnosis. Therefore, the identification of robust and easy-to-use biomarkers for the early identification of patients at high risk in this patient group is essential. Although not perfect, the clinical PET model is the best we have to select patients at high risk with limited additional costs and limited additional time because, on an average, MTV can be calculated for patients within 3 to 6 minutes, taking up to 10 to 20 minutes for complex cases.43
The focus of a validation study should not be on the statistical testing of differences in performance but on the generalizability of the model in other settings.44,45 A prediction model ideally provides valid predictions of outcomes for individual patients in real life. Our study showed that our clinical PET model was generalizable because it remained predictive of outcome in all external studies, which were clinical cohorts of unselected patients that can represent real-life settings. After updating the model (ie, recalibration of the intercept and coefficients), comparable model and diagnostic performances were confirmed. However, case-mix differences between individual studies were present regarding patient characteristics, outcome, treatment, and 18F-FDG–PET parameters. This led to different model performances between studies for both IPI and clinical PET model. This is most prominent in HOVON-130, a study with most aberrant patient and 18F-FDG–PET characteristics, compared with other studies, because it only included patients with MYC gene rearrangements, and a subgroup of these patients showed poor survival rates irrespective of disease burden quantified based on radiomics features.46 The SAKK study mainly included patients at low risk, which led to poor performance of the IPI risk score. However, our clinical PET model was still able to accurately predict the outcome for these patients at low risk. The patient characteristics in Table 1 show that the NCRI and SAKK studies included relatively more patients at limited stages, whereas the HOVON-130, HOVON-84, and GSTT15 studies included more patients at advanced stages. These differences were also visible in the IPI score. These case-mix differences are more pronounced when the sample sizes are relatively small, which is the case for the GSTT15, HOVON-130, IAEA, NCRI, and SAKK studies. The uncertainty of the model increases, leading to a large range of CIs,47 possibly explaining the large variation in model performance. Regardless of these case-mix differences, the model performances of the clinical PET model always outperformed those of the IPI model. This led to a more accurate selection of patients at high risk, as shown by the decrease of 10% (IPI, 61.4% vs 51.9% for clinical PET model) in the survival for the high-risk group and an increase of 14% (35.5 vs 49.1 respectively) for the PPV (compared with the IPI model).
Significant efforts have been made to standardize 18F-FDG–PET scanning, including initiatives by the European Association for Nuclear Medicine Research Limited and the US Society of Nuclear Medicine.48,49 However, the absence of a standardized methodology has hampered the use of quantitative PET parameters in clinical practice. However, multiple vendors of 18F-FDG–PET systems have implemented algorithms to calculate the MTV. Currently, dissemination features are included only in the context of the research. However, these features are relatively insensitive to differences in segmentation methods, acquisition, and reconstruction39,42 and are relatively simple to calculate. Therefore, implementation of the calculation of these radiomics features should be feasible in a reproducible manner in most clinical PET centers. We expect and hope that vendors will implement the calculation of radiomics features in their software in the foreseeable future, once more evidence on their clinical value becomes apparent. In the meantime, our image analysis tool, ACCURATE, is provided as an open tool to facilitate research use.
This study has several strengths. By applying 2 risk scores to the same individual patient data from high-quality studies, this analysis allowed for the direct comparison of risk indices. Furthermore, the applied PET quality control criteria and uniform analysis of the baseline 18F-FDG–PET/CT scans resulted in the inclusion of high-quality PET data. Moreover, survival data were harmonized by recalculating the follow-up between the original studies. We decided to truncate survival at 2 years because the most clinically relevant events occurred during this period. An individual patient data analysis reported that patients who are alive without progression at 2 years have similar survival rates as the age-, sex-, and country-matched population 7 years after this time.50 A limitation of our study was that for some patients included in the PETRA database, the baseline 18F-FDG–PET/CT scan was either not performed or performed on a PET-only system (235 out of 392). Therefore, not all patients were included in the post hoc analysis. However, we believe that for prospective trials, fewer patients will be excluded because of insufficient PET quality, given that there is increased awareness of scanning and anonymization procedures compared with the timeframe when prospective clinical trials were performed. Furthermore, we decided to include TTP as an outcome parameter, because PFS and overall survival are affected by aging.6 The outcome of older patients is determined not only by lymphoma but also by age-related comorbidities, adverse treatment effects, and limited life expectancy in general. Lastly, although most patients were treated with R-CHOP, differences in treatment regimens between studies existed with regard to the number of cycles and intensification of treatment.
In conclusion, the clinical PET model that was developed in the HOVON-84 data set remained predictive of outcome in 6 independent studies and had a better model performance than the currently used IPI risk score in all studies. Therefore, baseline 18F-FDG–PET radiomics features can be used to select patients at high risk more accurately than the IPI model, given its relatively higher model performance and PPV.
Acknowledgments
The authors thank all patients who participated in the trials and the collaborating investigators who kindly supplied their data. The authors also thank all data managers who collected the clinical data and 18F-FDG–PET/CT scans for individual studies.
This study was financially supported by the Dutch Cancer Society (VU 2018–11648). The PETAL trial was supported by grants from Deutsche Krebshilfe (107592 and 110515). S.F.B. acknowledges the support from the National Institute for Health and Care Research (RP-2-16-07-001). King’s College London and the UCL Comprehensive Cancer Imaging Centre are funded by the CRUK and EPSRC in association with the MRC and the Department of Health and Social Care (England). This work was also supported by core funding from the Wellcome/EPSRC Centre for Medical Engineering at King’s College London (WT203148/Z/16/Z) and the National Institute for Health and Care Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ National Health Service (NHS) Foundation Trust and King’s College London and the NIHR Clinical Research Facility.
The views expressed are those of the authors and not necessarily those of the NHS, NIHR, or the Department of Health and Social Care.
Authorship
Contribution: J.J.E., G.J.C.Z., O.S.H., H.C.W.d.V., R.B., and J.M.Z. contributed to the concept and design of this study; U.D., A.H., S.F.B., N.G.M., E.Z., T.G., P.J.L., and M.E.D.C. were responsible for data acquisition; J.J.E., G.J.C.Z., S.E.W., S.P., C.H., L.K., L.C., and S.C. performed PET/CT analyses; J.J.E. and M.W.H. performed statistical analyses; and all authors contributed to the interpretation of the data and all authors critically reviewed and approved the manuscript.
Conflict-of-interest disclosure: S.F.B. received departmental funding from Amgen, AstraZeneca, BMS, Novartis, Pfizer and Takeda. M.E.D.C. received financial support for the clinical trials from Celgene, BMS and Gilead. J.M.Z. received financial support for clinical trials from Roche, Gilead, and Takeda. The remaining authors declare no competing financial interests.
A complete list of the members of the PETRA Consortium appears in the supplemental Appendix.
Correspondence: J. J. Eertink, Department of Hematology, Amsterdam UMC, location VUmc, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands; e-mail: j.eertink@amsterdamumc.nl.
References
Author notes
All data are available on request from the corresponding author, J. J. Eertink (j.eertink@amsterdamumc.nl). Deidentified individual participant data can be requested through the PETRA consortium request platform at https://petralymphoma.org (petra@amsterdamumc.nl).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal