In this issue of Blood, Ansari-Pour et al1 present findings from their analysis on the largest whole-genome sequencing data set of relapsed and refractory multiple myeloma (rrMM), which comprised 418 tumor samples from 386 patients. The data were retrieved from 6 clinical trials that included patients refractory to lenalidomide and/or pomalidomide, which are 2 immunomodulatory imide drugs (IMiDs) that are commonly used therapeutics for newly diagnosed and relapsed patients with multiple myeloma (MM), respectively.
MM is characterized by remarkable inter- and intrapatient genomic heterogeneity.2,3 Recent analyses of large multiomics data from newly diagnosed patients with MM revealed a complex genomic landscape and the existence of multiple genetic subtypes with distinct and well-defined sets of co-occurring genetic alterations and transcriptomic features, which could stratify patients according to their risk of progression after first-line therapy and overall survival.4,5 Broadly, chromosomal translocations involving the immunoglobulin locus on chromosome 14 and oncogenes such as NSD2, CCND1, and MAF and hyperdiploidy, a genetic abnormality defined by the presence of 3 copies of at least 2 odd-numbered chromosomes, are often the main initiating events. Additional recurrent alterations, including both single nucleotide variants and copy number changes, can then associate with translocations and hyperdiploidy following different patterns of co-occurrence and mutual exclusivity, further contributing to increased genomic complexity. Longitudinal studies in patients with MM sequenced at diagnosis and subsequent relapses have revealed a diverse landscape of clonal and subclonal aberrations, enabling the identification of initiating driver events and alterations arising in later disease phases that drive relapse.5-7
Although recent studies have explored disease evolution in a small number of longitudinally profiled patients, the novel study by Ansari-Pour et al is the first to examine whole-genome data from a large population of patients refractory to a specific category of drugs (IMiDs) and compare the findings with a large data set from newly diagnosed, although unrelated, patients to identify the key features of rrMM. The high coverage breadth and uniformity of whole-genome sequencing allow the identification of broad events in MM tumors, such as whole-genome duplication and chromosomal translocations, which cannot be accurately detected using more targeted approaches, including whole-exome sequencing.
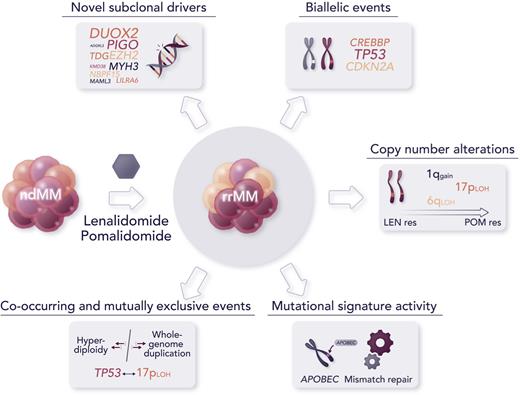
The major findings of this study include the discovery of novel candidate subclonal driver mutations in oncogenes and tumor suppressors, including the epigenetic regulator EZH2 and transcription coactivator MAML3, as well as noncoding somatic variants, such as the TP53 binding protein TP53BP1, undergoing significant clonal expansion or increased frequency in patients with rrMM than in newly diagnosed MM (ndMM). Some high-risk copy number alterations, such as 1q gain and 17p loss of heterozygosity, were not only enriched in patients with rrMM compared with patients with ndMM but were also significantly detected as co-occurring in some patients with rrMM, so called double-hit events, and showed an increasing trend in progressive IMiD resistance to lenalidomide and pomalidomide. Biallelic events, that is alterations affecting both alleles of a gene, were identified in driver genes at a higher prevalence in rrMM than in ndMM, with some events seemingly specific to rrMM, such as those involving CDKN2A and CREBBP, and biallelic inactivation of TP53, a high-risk abnormality associated with aggressive disease, whose frequency was almost twofold greater in the relapsed cohort than in the ndMM cohort. Furthermore, mutational signatures attributed to APOBEC (SBS2/SBS13) and defective mismatch repair (SBS12) were identified at increased activity from diagnosed to refractory stages, the latter not previously described in MM.8 Significant elevation of mutational burden, driven by more than a twofold increase in single nucleotide variants, was observed in patients with a higher defective mismatch repair signature activity. This suggests a possible association with subclones that expand under therapeutic selection pressure. As observed by the authors, subclonal complexity appeared to enrich with disease evolution rather than decrease with treatment pressure, as it would be reasonable to expect, which is in line with a recent report of emerging unique subclones after multiple relapses in patients evaluated longitudinally.6 This indicates that subclonal tumor evolution should be thoroughly investigated, possibly with single-cell methodologies, to better understand disease relapse and guide therapeutic choices.9
Although this novel study contributes to advancing our knowledge of the genomics of rrMM and provides the research community with an important data resource, it also comes with a few major caveats. First, despite its use of the largest data set of whole-genome sequencing in rrMM, the analysis was not powered to identify significantly recurring alterations, given the considerable interpatient heterogeneity of MM. Increasing the data set size would likely discover additional variants involved in therapy resistance and/or further consolidate current findings. Second, the comparison with patients with ndMM was done only at the population level using an independent data set. Sequential sampling of patients from diagnosis to relapse would provide a more accurate analysis of the clonal landscape and subclonal structure evolution, as demonstrated by previous recent studies, albeit in much smaller patient cohorts. Furthermore, the inclusion of transcriptomic data from RNA sequencing would provide an important complement to the analysis and allow us to contextualize the findings within more specific downstream molecular mechanisms and pathways. Finally, although the identification of novel driver events was performed using state-of-the-art computational tools and further evaluated by orthogonal analyses to establish their oncogene and tumor suppressor potential as well as their impact on MM cell lines via CRISPR screening, follow-up experiments and animal models will be necessary to determine that these events are, indeed, drivers of relapse and resistance to IMiDs (see figure).
Major findings from the novel study by Ansari-Pour et al include the discovery of novel abnormalities associated with refractoriness to IMiDs and significant changes in the recurrence of high-risk alterations from diagnosis to relapse. Professional illustration by Somersault18:24.
Major findings from the novel study by Ansari-Pour et al include the discovery of novel abnormalities associated with refractoriness to IMiDs and significant changes in the recurrence of high-risk alterations from diagnosis to relapse. Professional illustration by Somersault18:24.
Although a small fraction of patients with MM experiences a relapse-free interval of more than 10 years after the first therapy, suggesting complete eradication of the disease, most patients eventually relapse and face the challenges of an increasingly difficult disease management. Understanding how the MM genome evolves and becomes refractory to treatment is key to developing novel therapeutic strategies.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal