In this issue of Blood, Sharma et al1 demonstrate that gene transcription in macrophages contributes to the development of virus-associated coagulopathy in mice infected with the murine coronavirus MHVA59, which was used as a surrogate for pulmonary SARS-CoV-2 infection in this study. Because coagulopathy is a major complication of SARS-CoV-2 infection, this mechanism of regulation may be relevant to the ongoing pandemic. In Sharma’s study, transcriptional control is mediated through changes in chromatin structure wrought by the addition of a trimethyl group on the fourth lysine residue in the tail of histone 3 (H3K4Me3). Although many enzymes can set the H3K4Me3 mark, in this model the enzyme responsible is mixed lineage leukemia 1 (MLL1/KMT2a), which derives its name from gene rearrangements found in both lymphoblastic and myeloid leukemias. The team chose to assess the role of MLL1 in viral infection because previous studies have demonstrated its importance in macrophage activation (see figure).
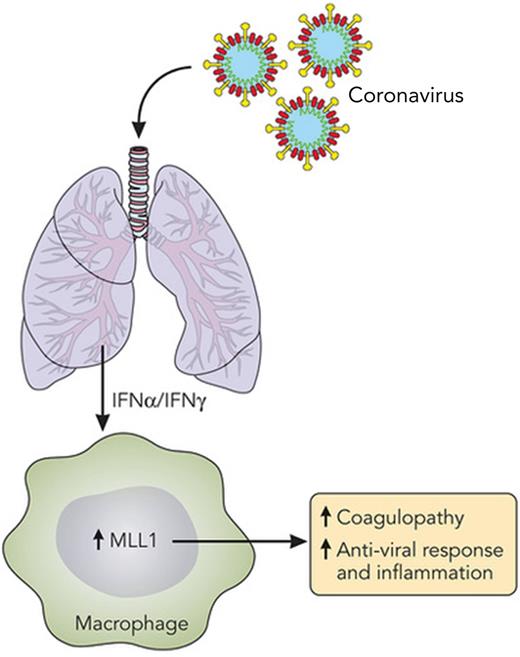
MLL1 is central to macrophage activation. A summary of the work by Sharma et al,1 depicting increased MLL1 expression in macrophages due to IFNs produced as a result of pulmonary coronavirus infection. MLL1 expression is a key factor in macrophage activation, causing coagulopathy and inflammation and inducing an antiviral response. Professional illustration by Patrick Lane, ScEYEnce Studios.
MLL1 is central to macrophage activation. A summary of the work by Sharma et al,1 depicting increased MLL1 expression in macrophages due to IFNs produced as a result of pulmonary coronavirus infection. MLL1 expression is a key factor in macrophage activation, causing coagulopathy and inflammation and inducing an antiviral response. Professional illustration by Patrick Lane, ScEYEnce Studios.
Sharma et al used a mouse model in which MLL1 was deleted using a Cre/Flox system, where the Cre recombinase is expressed under the control of Lyz2 gene promoter, resulting in MLL1 deletion in myeloid cells. They demonstrate that mice with MLL1-deficient macrophages are less prone to coagulopathy, with increased bleeding times and decreased expression of urokinase, the urokinase receptor, and factor III. They then confirm their results in vitro through small interfering RNA (siRNA) knockdown of MLL1 in macrophages. Given the essential role of MLL1 in hematopoietic cell expansion,2 the results of the siRNA experiments are critical to demonstrate that the observed phenotype is not due to alterations in macrophage development. Instead, inhibition of MLL1 just prior to macrophage stimulation reduces the transcription of genes related to coagulopathy. Finally, the team replicated their findings in humans. Their data demonstrate an increase in expression of MLL1 and coagulopathy factors in peripheral blood monocytes from hospitalized patients with COVID-19 when compared with monocytes of hospitalized patients that were not positive for COVID-19.
In concurrence with previous research, the team identifies interferon α (IFN-α) and IFN-γ as potential cytokines that augment MLL1 expression. From the prior study, we know that MLL1 increases STAT4 expression in macrophages.3 STAT4 induces an antiviral program that includes expression of IFN-β, tumor necrosis factor, and interleukin-6 as well as stabilization of the protein produced by retinoic acid-inducible gene 1 (RIG-I).4 When taken together with the recent work by Sharma et al, these results outline a role for macrophage-expressed MLL1 in the antiviral response, which includes coagulopathy and inflammation. Because of the variety of MLL1-dependent genes identified in macrophages/monocytes, there are 2 possible interpretations of the data: the first is that MLL1 deletion specifically reduces the coagulopathy response in macrophages; the second is that MLL1 is a general regulator of transcription, including coagulopathy, during macrophage activation.
Four additional studies that define the role of MLL1 in the context of wound healing portray MLL1 as a protein that is generally associated with inflammation. The first study examines the wound healing process and expression of MLL1 in the context of long-term postseptic immunosuppression.5 In this study, we observed delayed wound closure in mice that had recovered from sepsis, which was correlated with reduced MLL1 expression. We also observed reduced expression of MLL1 in the peripheral blood of postseptic patients for up to 3 years after their septic event. Two additional studies demonstrate that MLL1 promotes expression of the COX2 and Toll-like receptor (TLR) signaling pathways in macrophages during the wound healing process,6,7 with a fourth study showing that MLL1 expression is increased in peripheral blood monocytes from patients with diabetes, when hyperinflammatory conditions persist.8 These results suggest that increased MLL1 expression is not specific to viral infection. Although Sharma et al show a good correlation between coagulopathy and MLL1 expression in hospitalized patients diagnosed with SARS-CoV-2, this increase in MLL1 expression may only be indicative of the activated state of the macrophage. Further study of MLL1 expression that capitalizes on the heterogenous response to SARS-CoV-2 infection would clarify these results.
Although MLL1 may serve as a biomarker of inflammation, inhibiting the activity of MLL1 as a treatment for inflammation-based disease may be challenging. There are many MLL1 inhibitors under development9 directed toward treatment of leukemias with an MLL1 gene rearrangement. Some of these inhibitors will also target MLL1 in nonleukemic cells but are likely to cause unwanted side effects. Given the number of cells that express MLL1, the dose and timing of inhibition may be difficult to determine. If an anti-inflammatory therapy were to be developed that involved MLL1 inhibition, it would be optimal only for treatment of an acute disease state, which would preserve hematopoietic cell development in the long term. Another approach may be to target the methyltransferase domain of MLL1, leaving the rest of the protein intact, which would preserve the self-renewal capacity of adult hematopoietic stem cells.10
Even if targeting MLL1 itself is not feasible, the work by Sharma et al hints at the possibility of developing novel therapies for coagulopathy and acute inflammation. The team’s work highlights the precise control of transcriptional processes that are critical components of macrophage activation. Collectively, the work focused on MLL1 demonstrates that removing a single enzyme in 1 cell type is sufficient to change the phenotype in 2 different models of inflammation (viral infection and wound healing) and reduce transcripts related to unique inflammatory pathways including coagulopathy, antiviral responses, TLR signaling, and prostaglandin production. However, MLL1 is only 1 of over a dozen enzymes that can add a methyl group to lysine 4 of histone 3. Furthermore, methylation is only 1 of 9 histone modifications that can be attached to the amino acid residues in histone tails, resulting in changes in transcriptional rate. Finally, to accomplish the H4K4Me3 modification, MLL1 acts in concert with a dynamic group of adaptor and scaffold proteins and enzymes to modify multiple histone tails simultaneously. The complexity of this system, although daunting, provides a myriad of possibilities for targeting the transcriptional control of inflammatory processes. The challenge will be to identify those enzymes and histone modifications that are dispensable for cells at rest but essential for inflammation.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal