Page 2202: In the article title, the phrase “tumor-immune microenvironment” should not have a hyphen.
Page 2205: The first sentence under “CD40L-mediated in vitro activation of CD70 expression in MCL cell lines” should read “Stable transduced Z138 SOX11-knockdown (Z138SOX11KD), JVM2 ectopically overexpressing SOX11 (JVM2SOX11+), and their respective control (Z138CT and JVM2CT) MCL cell line models, previously generated by our group,8,12 were used for in vitro experiments.”
Page 2209: In the first paragraph under “SOX11-mediated CD70 expression in MCL cells is activated by CD40L in vitro,” the sentence that begins “ChIP-qPCR” should read “ChIP-qPCR experiments showed fold enrichment of the CD70 locus in the ChIPs of SOX11+ cell line (Z138 wild type [WT] and JVM2SOX11+12 and Z138CT [red]) DNAs over their inputs, with a highly significant enrichment compared with non-SOX11-expressing cell lines (JVM2CT and Z138SOX11KD8 [blue]) (Figure 5A-B).”
There are corrections in some figures due to errors in figure processing during the publication process.
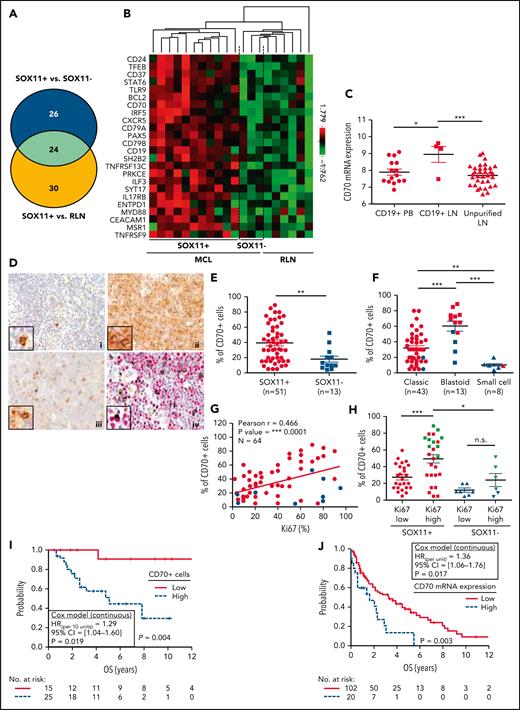
CD70 overexpression in nodal MCLs. (A) Venn diagram showing the overlap between the NanoString-based statistically significant upregulated genes in SOX11+ compared with SOX11− nodal MCLs (50 genes; blue circle) and SOX11+ compared with RLNs (54 genes; yellow circle). (B) Heatmap showing the scaled expression levels of common 24 significant upregulated genes in SOX11+ compared with SOX11− nodal MCLs and RLNs. Red represents increased expression and green reduced expression. Genes with an adjusted P value (Q-value) < .15 were considered. (C) CD70 mRNA expression levels in unpurified lymph node MCL samples (unpurified LN; n = 34), CD19+ purified cells from LN samples (CD19+ LN; n = 4) and peripheral blood samples (CD19+ PB; n = 15) (GSE70910). ∗q value < .05, ∗∗∗q value < .001. (D) Representative histological sections from (i) an RLN sample, (ii) SOX11+, and (iii) SOX11− nodal MCL biopsy, stained with specific anti-human CD70 antibody (×100). A double IHC staining with anti-human CD70 (brown) and (iv) anti-human cyclin D1 (red) antibodies (×100) was performed in the same SOX11− nodal MCL case as in (iii) to confirm the expression of CD70 by tumor cells (black arrows). Pictures contain insets with magnification (×400). (E) IHC quantifications of CD70+ cells in our series of SOX11+ (n = 51) and SOX11− (n = 13) nodal MCL primary samples. (F) IHC quantification of CD70 expression in nodal samples according to classic, blastoid/pleomorphic, or small cell MCL variants. (G) Positive correlation between CD70+ and Ki67+ cells, quantified by IHC in our series of nodal MCL. SOX11+ MCL are indicated in red, whereas SOX11− MCLs are in blue. Graphs show Pearson correlation coefficient (r), P value, and number of cases analyzed (N). (H) IHC quantification of CD70 expression in nodal MCL samples according to high (Ki67 >30%) and low (Ki67 ≤30%) proliferation rates in our series of SOX11+ (red) and SOX11− (blue) primary MCL cases. MCL cases diagnosed as blastoid/pleomorphic cytological variant are highlighted in green. The significance of differences was determined by independent samples Student t test: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. (I) Kaplan-Meier curve and Cox regression showing the association of CD70+ cells, quantified by IHC using our series of SOX11+ nodal MCL primary samples (n = 40), with OS. (J) Kaplan-Meier curve and Cox regression showing the association of CD70 mRNA expression with OS, using previously published GEP from nodal samples and clinical data from 122 nodal SOX11+ MCL primary cases (GSE93291). High values were defined by Maxstat (cutoff point IHC = 25%, cutoff point mRNA = 9.9). Log-rank test P values, hazard ratios (HR) with 95% confidence interval (CI), and Cox regression P values are shown.
CD70 overexpression in nodal MCLs. (A) Venn diagram showing the overlap between the NanoString-based statistically significant upregulated genes in SOX11+ compared with SOX11− nodal MCLs (50 genes; blue circle) and SOX11+ compared with RLNs (54 genes; yellow circle). (B) Heatmap showing the scaled expression levels of common 24 significant upregulated genes in SOX11+ compared with SOX11− nodal MCLs and RLNs. Red represents increased expression and green reduced expression. Genes with an adjusted P value (Q-value) < .15 were considered. (C) CD70 mRNA expression levels in unpurified lymph node MCL samples (unpurified LN; n = 34), CD19+ purified cells from LN samples (CD19+ LN; n = 4) and peripheral blood samples (CD19+ PB; n = 15) (GSE70910). ∗q value < .05, ∗∗∗q value < .001. (D) Representative histological sections from (i) an RLN sample, (ii) SOX11+, and (iii) SOX11− nodal MCL biopsy, stained with specific anti-human CD70 antibody (×100). A double IHC staining with anti-human CD70 (brown) and (iv) anti-human cyclin D1 (red) antibodies (×100) was performed in the same SOX11− nodal MCL case as in (iii) to confirm the expression of CD70 by tumor cells (black arrows). Pictures contain insets with magnification (×400). (E) IHC quantifications of CD70+ cells in our series of SOX11+ (n = 51) and SOX11− (n = 13) nodal MCL primary samples. (F) IHC quantification of CD70 expression in nodal samples according to classic, blastoid/pleomorphic, or small cell MCL variants. (G) Positive correlation between CD70+ and Ki67+ cells, quantified by IHC in our series of nodal MCL. SOX11+ MCL are indicated in red, whereas SOX11− MCLs are in blue. Graphs show Pearson correlation coefficient (r), P value, and number of cases analyzed (N). (H) IHC quantification of CD70 expression in nodal MCL samples according to high (Ki67 >30%) and low (Ki67 ≤30%) proliferation rates in our series of SOX11+ (red) and SOX11− (blue) primary MCL cases. MCL cases diagnosed as blastoid/pleomorphic cytological variant are highlighted in green. The significance of differences was determined by independent samples Student t test: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. (I) Kaplan-Meier curve and Cox regression showing the association of CD70+ cells, quantified by IHC using our series of SOX11+ nodal MCL primary samples (n = 40), with OS. (J) Kaplan-Meier curve and Cox regression showing the association of CD70 mRNA expression with OS, using previously published GEP from nodal samples and clinical data from 122 nodal SOX11+ MCL primary cases (GSE93291). High values were defined by Maxstat (cutoff point IHC = 25%, cutoff point mRNA = 9.9). Log-rank test P values, hazard ratios (HR) with 95% confidence interval (CI), and Cox regression P values are shown.
SOX11-dependent CD70 expression is induced by CD40L in vitro in MCL cells. ChIP-qPCR experiments showing the enrichment of specific CD70 loci (1393-1119 bp upstream of the transcription start site), identified by SOX11 specific ChIP-chip experiments in (A) 2 SOX11+ MCL cell lines, Z138 wild-type (Z138WT) and JVM2SOX11+, and its control SOX11− MCL cell line (JVM2CT)12 and (B) in Z138 SOX11-knockdown (Z138SOX11KD) and its SOX11+ control (Z138CT) MCL cell line,8 were used for SOX11-specific ChIP-qPCRs experiments. DNA enrichment is displayed as fold change relative to its respective input chromatin and JVM2CT and Z138SOX11KD enrichment, respectively. (C) RT-qPCR quantification of CD70 mRNA levels (D) and FC quantification of CD70 protein expression levels analyzed in our stable transduced cell lines Z138CT/Z138SOX11KD incubated with vehicle (phosphate-buffered saline [PBS]) or 50 ng/mL CD40L, for 6 hours. Results are represented as fold change in CD40L-treated cells relative to PBS-treated cells. The significance of difference was determined by independent samples Student t test: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
SOX11-dependent CD70 expression is induced by CD40L in vitro in MCL cells. ChIP-qPCR experiments showing the enrichment of specific CD70 loci (1393-1119 bp upstream of the transcription start site), identified by SOX11 specific ChIP-chip experiments in (A) 2 SOX11+ MCL cell lines, Z138 wild-type (Z138WT) and JVM2SOX11+, and its control SOX11− MCL cell line (JVM2CT)12 and (B) in Z138 SOX11-knockdown (Z138SOX11KD) and its SOX11+ control (Z138CT) MCL cell line,8 were used for SOX11-specific ChIP-qPCRs experiments. DNA enrichment is displayed as fold change relative to its respective input chromatin and JVM2CT and Z138SOX11KD enrichment, respectively. (C) RT-qPCR quantification of CD70 mRNA levels (D) and FC quantification of CD70 protein expression levels analyzed in our stable transduced cell lines Z138CT/Z138SOX11KD incubated with vehicle (phosphate-buffered saline [PBS]) or 50 ng/mL CD40L, for 6 hours. Results are represented as fold change in CD40L-treated cells relative to PBS-treated cells. The significance of difference was determined by independent samples Student t test: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
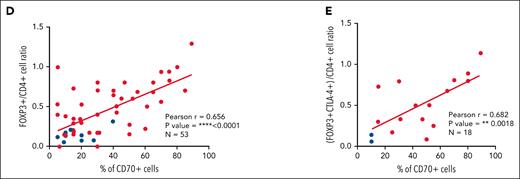
High number of intratumoral Treg cells correlates with high numbers of CD70+ cells in SOX11+ nodal MCLs, and it is associated with shorter OS of MCL patients. (D-E) Positive correlation between CD70+ cells and FOXP3+/CD4+ T-cell ratio (D) and CD70+ cells and (FOXP3+CTLA4+)/CD4+ T-cell ratio (E) in SOX11+ (red) and SOX11− (blue) nodal MCLs, quantified by IHC in our series of nodal MCLs. Graphs show Pearson correlation coefficient (r), P value, and number of cases analyzed (N).
High number of intratumoral Treg cells correlates with high numbers of CD70+ cells in SOX11+ nodal MCLs, and it is associated with shorter OS of MCL patients. (D-E) Positive correlation between CD70+ cells and FOXP3+/CD4+ T-cell ratio (D) and CD70+ cells and (FOXP3+CTLA4+)/CD4+ T-cell ratio (E) in SOX11+ (red) and SOX11− (blue) nodal MCLs, quantified by IHC in our series of nodal MCLs. Graphs show Pearson correlation coefficient (r), P value, and number of cases analyzed (N).
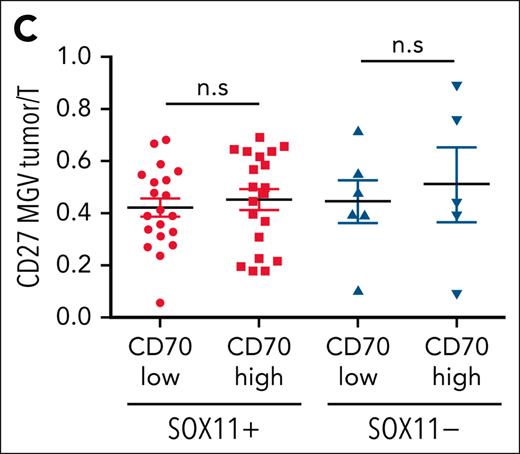
CD27 in MCLs. (C) Mean gray value (MGV) quantification of CD27 staining (see supplemental Methods) in cases with low and high CD70 expression.
CD27 in MCLs. (C) Mean gray value (MGV) quantification of CD27 staining (see supplemental Methods) in cases with low and high CD70 expression.
The publisher apologizes for these errors, which (except for changing “pb” to “bp” in the legend to Figure 5) have been corrected in the online version of the article.

![SOX11-dependent CD70 expression is induced by CD40L in vitro in MCL cells. ChIP-qPCR experiments showing the enrichment of specific CD70 loci (1393-1119 bp upstream of the transcription start site), identified by SOX11 specific ChIP-chip experiments in (A) 2 SOX11+ MCL cell lines, Z138 wild-type (Z138WT) and JVM2SOX11+, and its control SOX11− MCL cell line (JVM2CT)12 and (B) in Z138 SOX11-knockdown (Z138SOX11KD) and its SOX11+ control (Z138CT) MCL cell line,8 were used for SOX11-specific ChIP-qPCRs experiments. DNA enrichment is displayed as fold change relative to its respective input chromatin and JVM2CT and Z138SOX11KD enrichment, respectively. (C) RT-qPCR quantification of CD70 mRNA levels (D) and FC quantification of CD70 protein expression levels analyzed in our stable transduced cell lines Z138CT/Z138SOX11KD incubated with vehicle (phosphate-buffered saline [PBS]) or 50 ng/mL CD40L, for 6 hours. Results are represented as fold change in CD40L-treated cells relative to PBS-treated cells. The significance of difference was determined by independent samples Student t test: ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/142/13/10.1182_blood.2023021334/2/m_blood_bld-2023-021334-gr2.jpeg?Expires=1765919398&Signature=Nu4rXGPaJhf9RAVTQuWDmEkUE6hY-pyu2tdkXURf6c2yBSBRAQdCfo7LDLvKqpGT2taIoVnqVgx~vAtUkJS82~626uvPW7Nx7cBO5sWnDa7o9HlWsGn8lFa2~dwxO22aIF5b40iJHAdRjUfflHaNySgmpjOaEcX0iF~i6ne4CrlUu9X69CITjOXW9199XjtdVkIuYKKhrTyJmfsrfjlybfALuPvqZ2cMOzTEyb5~u962eyrCdD6cNGZx4CJBCLZgQu7q7Wu4Q9N8SX63b3bGofFtS5TXCBZNDkLOZ37mE92YGAX2R2n5xYstI2Dd4h0ifmZncYdcSveYO8dBnwRrwQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal