Introduction
Acute myeloid leukemia (AML) and myelodysplastic neoplasm (MDS) are common hematological malignancies, and the majority of patients carry genetic mutations, structural variants (SVs) and copy number variants (CNVs). Identifying these variants is important for diagnosis and prognosis, and in some cases can alter treatment management and outcomes. Methods for analyzing SVs and CNVs include karyotype analysis, fluorescence in situ hybridization, gene chips, and high-throughput gene sequencing technologies. Optical genome mapping (OGM) is a novel ultra-long single-molecule DNA detection based high throughput SVs analysis technology, which has the characteristics of whole genome coverage, high detection incidence and sensitivity. There were few publications presenting technical concordance of SVs detection between karyotyping and OGM, and there are no systematic studies comparing the concordance of OGM versus transcriptome sequencing (RNA-seq) in detecting gene fusion. This study aimed to compare the concordance of OGM and RNA-seq in the detection of fusion genes in AML and MDS cases, and the additional findings using OGM technology.
Methods
Bone marrow aspirates of 52 newly diagnosed or relapsed/refractory AML/MDS patients were recruited for OGM, RNA-seq, and leukemia-related gene DNA panel sequencing. Among these cases, 36 males and 16 were females, 15 pediatric (< 18 years) and 37 adults (≥18 years), with a median age of 31 years.
Results
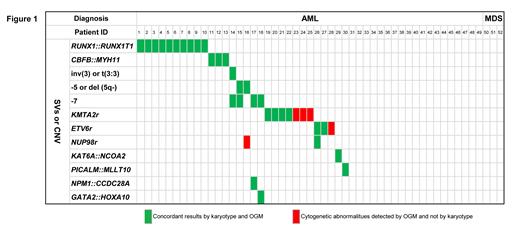
Based on the RNA-seq results, 29/52 (55.8%) cases were detected with pathogenic gene fusions, including RUNX1::RUNX1T1, CBFB::MYH11, KMT2Aretc. (Figure 1). All those fusions were detected by OGM. For SVs with VAF>5%, the concordance was 98.1% (51/52 cases) when comparing OGM with karyotyping. OGM missed one case whose translocation happened close to the centromere. OGM reported more pathological variants with clinically significant in 5/52 cases (9.62%) and helped revise karyotype results in 1 case with clarifying marker chromosome. According to the ELN-2022 risk stratification mainly based on karyotype results, OGM's report results helped revise the risk stratification of 3 patients from intermediate to high risk. Fifty of 52 (96.2%) patients carried the gene mutation detected by the gene sequencing panel (Figure 1). In the samples of 4 patients who relapsed after receiving allogeneic hematopoietic stem cell transplantation (allo-HSCT) with donors and patients of different genders, we can infer the proportion of male and female DNA based on the copy number of chr Y detected by OGM, and then determine the proportion of donor cells.
Conclusion
This is the first AML cohort study to systematically compare the performance of OGM and RNA-seq, and the reported results of fusion genes are completely consistent. This is also the first report of applying the OGM technology in patients who relapsed after allo-HSCT and for chimerism analysis. Combined using OGM and targeted sequencing to fully characterize the CNV, SV, and mutation can be used for accurate stratification risk of AML/MDS. In conclusion, combining OGM and NGS can significantly improve the detection rate of genetic abnormalities with important pathological/diagnostic significance, and may bring new discoveries on SVs and pathogenic mechanisms.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal