In this issue of Blood, Aoki et al report that the canonical BAF (cBAF) complex is required for the migration and survival of the malignant T cells in T-cell acute lymphoblastic leukemia (T-ALL).1
Recently, a number of studies have dissected important regulatory roles for the mammalian switch/sucrose nonfermentable (SWI/SNF) complex in the activation, memory, and exhaustion of T cells.2-6 The SWI/SNF complex is 1 of 4 adenosine triphosphate (ATP)-dependent chromatin remodeling complexes in mammalian cells along with the imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), and inositol auxotrophy 80 (INO80) complexes. SWI/SNF exists in 3 forms: cBAF, polybromo-associated BAF (PBAF), and noncanonical BAF (ncBAF), each with distinct subunits and unique roles in gene expression regulation. SWI/SNF complexes reposition nucleosomes in an ATP-dependent manner to allow transcription factors to access DNA to modify gene expression and perform cell type–specific gene regulatory activities. As such, the aforementioned studies previously identified unique roles and requirements for each form of the SWI/SNF complex at different stages of normal T-cell function. Now, Aoki et al have made the exciting discovery that in T-ALL, cBAF, but not PBAF or ncBAF, is uniquely required for migration and survival of the malignant leukemic cells. Given the development of SWI/SNF targeted therapeutics, these findings have direct implications for future treatments of T-ALL.
Aoki et al performed a genetic screen to identify the factors required for T-ALL cells to migrate toward the chemokine CXCL12. Interaction of CXCR4 on the surface of T-ALL cells with its ligand CXCL12 in the T-ALL microenvironment is critical for T-ALL cell survival.7 Interestingly, their whole genome CRISPR screen revealed CXCR4 as the top hit, as expected, followed by several components of the cBAF complex. Follow-up validation studies tested the requirement of cBAF vs PBAF and ncBAF in migration of human T-ALL cell lines toward CXCL12. These studies confirmed the initial genome-wide CRISPR screen result and revealed that cBAF specifically is required in T-ALL cell migration.
The result described is particularly intriguing from a therapeutic perspective as chemical inhibitors of the ATPases required for SWI/SNF function are now in early clinical development for a variety of cancers, including acute myeloid leukemia (AML).8 Moreover, chemical degraders of SWI/SNF, based on the inhibitors, have also been developed.9 Importantly, application of these chemical inhibitors and degraders of cBAF resulted in the same impairment of T-ALL cell migration as seen with genetic knockout of cBAF members. Furthermore, beyond simply inhibiting T-ALL migration, genetic or pharmacologic inhibition of cBAF also resulted in decreased fitness of T-ALL cells.
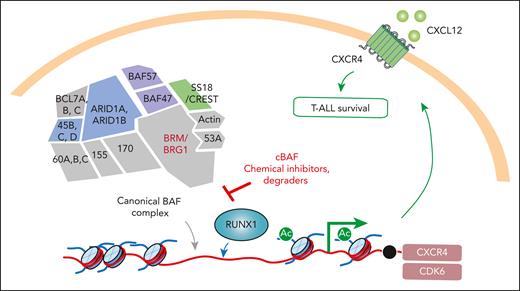
To explore why cBAF is required in the survival of T-ALL cells, the investigators evaluated chromatin accessibility upon cBAF depletion. Prior studies have revealed that the gene regulatory activities of SWI/SNF are controlled by the repertoire of transcription factors expressed in that cell type.2 Consistent with this concept, Aoki et al identified that regions of the genome where chromatin accessibility was altered upon cBAF loss overlapped with those bound by the transcription factor RUNX1 (see figure). The authors were able to surmise that cBAF loss resulted in eviction of RUNX1 from the genome, which caused downregulation of RUNX1-regulated genes, including CXCR4 as well as CDK6. This change in cBAF and RUNX1 gene regulatory activity was dynamic as restoration of cBAF complex expression rescued the expression of RUNX1-regulated genes.
The cBAF complex is required for migration and survival of T-ALL. In a genome-wide CRISPR screen aimed at identifying regulators of migration of T-ALL cells toward CXCL12, Aoki et al discovered that the ATP-dependent chromatin remodeling cBAF complex is required for CXCR4 expression, CXCL12 migration, and T-ALL survival. cBAF regulates chromatin in T-ALL cells in a manner that allows for the transcription factor RUNX1 to access promoter and drive expression of CXCR4 and CDK6. As such, small molecule inhibitors and degraders of cBAF result in death of T-ALL cells.
The cBAF complex is required for migration and survival of T-ALL. In a genome-wide CRISPR screen aimed at identifying regulators of migration of T-ALL cells toward CXCL12, Aoki et al discovered that the ATP-dependent chromatin remodeling cBAF complex is required for CXCR4 expression, CXCL12 migration, and T-ALL survival. cBAF regulates chromatin in T-ALL cells in a manner that allows for the transcription factor RUNX1 to access promoter and drive expression of CXCR4 and CDK6. As such, small molecule inhibitors and degraders of cBAF result in death of T-ALL cells.
Overall, this study expands our understanding of the gene regulatory machine of T cells spanning from activation, memory, exhaustion, and now transformation. Moreover, this study identifies cBAF as a pharmacologic target in the treatment of T-ALL and supports prior work highlighting CXCR47 and CDK610 as therapeutic targets in the disease.
Several major questions regarding the therapeutic implications of this work will be important to evaluate moving forward. First, Aoki et al identified that cBAF regulates T-ALL migration and survival in a manner dependent on RUNX1. However, there are genetic subsets of T-ALL where RUNX1 is mutated and altered in its transcriptional function. It is not clear if the requirement for cBAF is consistent across subtypes of T-ALL with distinct RUNX1 genotypes and expression. More broadly, it is known that the ATPase subunits of cBAF and PBAF are required for normal hematopoiesis and immune cell function. The therapeutic index of chemical inhibition and degradation of these complexes is currently unknown in patients. The ongoing early phase clinical evaluation of these agents in AML and solid tumors will hopefully address this issue and motivate translation of the work by Aoki et al to patients with residual and refractory T-ALL in need of new treatments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal