Emicizumab improves the procoagulant activity of select loss-of-function factor IX (FIX) variants with likely dysfunctional assembly of the intrinsic Xase complex, resulting in hemophilia B (HB). FVIII mimetics may represent an alternative nonfactor therapy for select patients with HB.
TO THE EDITOR:
Hemophilia A (HA) and hemophilia B (HB) are X-linked bleeding disorders that are caused by a reduction in factor VIII (FVIII) or factor IX (FIX) activity, respectively.1,2 Activated FVIII (FVIIIa) binds FIXa on anionic membranes to form the intrinsic Xase enzyme complex, which activates factor X (FX).3
HA and HB traditionally have been treated with intravenous factor replacement.1,2 Recently, HA management has been revolutionized with the advent of emicizumab, a bispecific antibody that can bind FIXa and FX to allow the former to cleave the latter without FVIIIa.4-6 Emicizumab is administered subcutaneously and provides highly efficacious prophylaxis for patients with HA.5-7 These attributes have led to rapid uptake of emicizumab for severe HA, especially among pediatric patients. Recent results support its use in nonsevere HA7 and other FVIII-deficient disorders, such as severe von Willebrand disease.8,9
In contrast, prophylaxis for HB still requires intravenous infusions of FIX. Subcutaneously administered nonfactor therapies (NFTs) are being evaluated in HB, but clinical development has been disrupted by thrombotic complications.10 There are no current NFTs in development that specifically target the deficiency in FIX activity. Previous personalized medicine approaches for HB using drug-induced readthrough identified a limited number of susceptible F9 genotypes.11,12
Although described as an FVIII mimetic, the emicizumab-FIXa complex is biochemically distinct from FVIIIa-FIXa.13,14 We hypothesized that these differences can be leveraged to repurpose emicizumab as a potential treatment for certain HB F9 genotypes. FIX variants with loss of function (LOF) caused by dysfunctional assembly of the intrinsic Xase enzyme complex (such as disrupted FVIIIa interactions) may be amenable to procoagulant rescue by FVIII mimetics. To test this hypothesis, we screened 105 HB-causing15 FIX variants; we found 41 variants at 23 amino acid positions that demonstrated improved procoagulant activity with emicizumab (Figure 1; supplemental Table 1, available on the Blood website). We confirmed the translational relevance of these results by similarly demonstrating improved procoagulant activity in select patient samples.
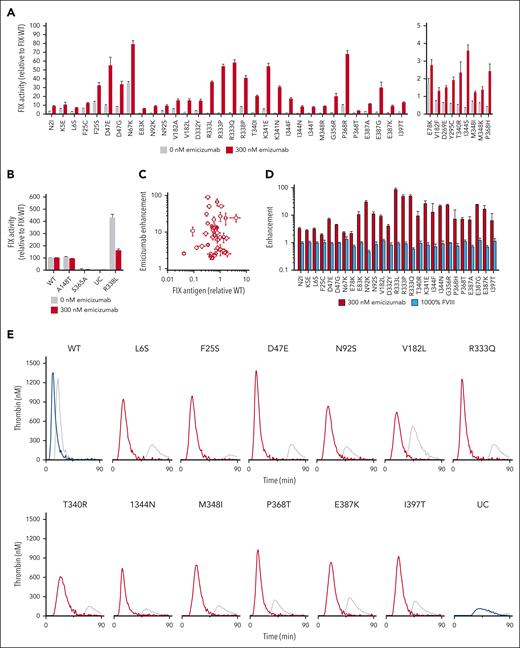
Evaluation of procoagulant enhancement of HB-causing FIX variants by emicizumab. (A) The FIX activity of conditioned media from HEK293 cells that transiently expressed recombinant select HB-causing FIX variants with 0 (gray) or 300 nM (red) emicizumab. Only variants with a greater than or equal to twofold enhancement with emicizumab are shown; nonrescuable variants are listed in supplemental Table 1. For clarity, variants with FIX activity with emicizumab >5% of normal are shown on the left, whereas variants with FIX activity with emicizumab 1% to 5% of normal are shown on the right. FIX activity was determined using an activated partial thromboplastin time (aPTT)–based assay with standard curves for 0 or 300 nM emicizumab (supplemental Figure 2). Values are normalized to recombinant FIX-WT activity expressed in the same experiment. Bars represent the mean ± standard error of mean of ≥9 independent transfections. (B) FIX activity of conditioned media with recombinant FIX protein controls with 0 (gray) or 300 nM emicizumab (blue). (C) Enhancement with emicizumab as a function of recombinant FIX antigen. (D) Enhancement with 300 nM emicizumab (red) or 1000% normal FVIII (light blue) of select HB-causing FIX variants. (E) Thrombograms of FIX variants with 0 nM (gray) or 300 nM emicizumab (red). Thrombograms of controls FIX-WT and UC with 0 nM (gray) or 300 nM emicizumab (blue). Coagulation was triggered with FXIa. Results are representative of ≥2 thrombograms from ≥2 independent transfections. Additional thrombograms of rescuable HB-causing variants are provided in supplemental Figure 3. (F) An aPTT-based clotting time of plasma from patients with moderate HB with FIX-I397T variant (patient 1) with increasing amounts of emicizumab in comparison with commercial severe HB plasma with undetectable FIX antigen (cross reactive material [CRM] negative). Points indicate the mean ± standard deviation of 4 measurements. The horizontal gray line represents the clotting time of normal human plasma. (G) Rotational thromboelastometry (ROTEM) thromboelastograms of whole blood from a patient with moderate HB with an FIX-I397T variant (patient 2) with 300 nM emicizumab or 100% recombinant FIX added ex vivo. Results are representative of 2 measurements. UC, unconditioned media.
Evaluation of procoagulant enhancement of HB-causing FIX variants by emicizumab. (A) The FIX activity of conditioned media from HEK293 cells that transiently expressed recombinant select HB-causing FIX variants with 0 (gray) or 300 nM (red) emicizumab. Only variants with a greater than or equal to twofold enhancement with emicizumab are shown; nonrescuable variants are listed in supplemental Table 1. For clarity, variants with FIX activity with emicizumab >5% of normal are shown on the left, whereas variants with FIX activity with emicizumab 1% to 5% of normal are shown on the right. FIX activity was determined using an activated partial thromboplastin time (aPTT)–based assay with standard curves for 0 or 300 nM emicizumab (supplemental Figure 2). Values are normalized to recombinant FIX-WT activity expressed in the same experiment. Bars represent the mean ± standard error of mean of ≥9 independent transfections. (B) FIX activity of conditioned media with recombinant FIX protein controls with 0 (gray) or 300 nM emicizumab (blue). (C) Enhancement with emicizumab as a function of recombinant FIX antigen. (D) Enhancement with 300 nM emicizumab (red) or 1000% normal FVIII (light blue) of select HB-causing FIX variants. (E) Thrombograms of FIX variants with 0 nM (gray) or 300 nM emicizumab (red). Thrombograms of controls FIX-WT and UC with 0 nM (gray) or 300 nM emicizumab (blue). Coagulation was triggered with FXIa. Results are representative of ≥2 thrombograms from ≥2 independent transfections. Additional thrombograms of rescuable HB-causing variants are provided in supplemental Figure 3. (F) An aPTT-based clotting time of plasma from patients with moderate HB with FIX-I397T variant (patient 1) with increasing amounts of emicizumab in comparison with commercial severe HB plasma with undetectable FIX antigen (cross reactive material [CRM] negative). Points indicate the mean ± standard deviation of 4 measurements. The horizontal gray line represents the clotting time of normal human plasma. (G) Rotational thromboelastometry (ROTEM) thromboelastograms of whole blood from a patient with moderate HB with an FIX-I397T variant (patient 2) with 300 nM emicizumab or 100% recombinant FIX added ex vivo. Results are representative of 2 measurements. UC, unconditioned media.
As detailed in the supplemental Methods, FIX protein was initially transiently expressed in mammalian cell culture and the conditioned media was assayed. Activity was measured using an activated partial thromboplastin time assay as done previously.16 The enhancement of FIX activity was defined as the ratio of FIX activity with and without emicizumab relative to wild-type FIX activity (FIX-WT). The thrombin generation assay (TGA) used for FIX-variant proteins and rotational thromboelastometry of whole blood of patients with HB are also described in the supplemental Methods. TGA enhancement is defined as the peak thrombin with and without emicizumab relative to FIX-WT corrected for thrombin generated from unconditioned media. FIX amino acid positions are numbered using the legacy system (initial methionine −45).16,17
The institutional review boards of the Children’s Hospital of Philadelphia (08-7008) and the University of Pennsylvania (833988) approved these studies.
To test the ability of FVIII mimetics, such as emicizumab, to rescue HB-causing LOF FIX variants, we transiently expressed recombinant FIX variants with amino acid substitutions at the positions implicated in the assembly of the intrinsic Xase complex,17-20 as well as controls. The former include positions within the putative FVIIIa-binding sites (supplemental Figure 1A) and those involved in the allosteric activation of FIXa upon FVIIIa binding (supplemental Figure 1B).17-20 FIX-variant expression in human embryonic kidney (HEK)293 cells qualitatively reflects the FIX antigen levels in clinical descriptions.16Figure 1A shows the FIX clotting activity of the rescuable LOF FIX variants in the absence or presence of a therapeutic amount of emicizumab (300 nM) in comparison with FIX-WT with the same amount of emicizumab. Although emicizumab shifts the curve relating clotting time to FIX activity to the left, the standard curves with and without emicizumab are approximately parallel (supplemental Figure 2). Thus, the relative increase in activity observed for these FIX variants in the presence of emicizumab is indicative that the FIX activity of these variants is specifically rescuable by FVIII mimetics rather than by the general procoagulant effect of emicizumab.
Rescuable LOF FIX variants are shown in Table 1. FIX variants that were evaluated but that did not demonstrate increased FIX activity with emicizumab are shown in supplemental Table 1. Figure 1B includes FIX variants that were not expected to be enhanced by emicizumab, including the nondeleterious Malmo variant FIX-A148T with activity and antigen levels similar to FIX-WT,21 the catalytically inert FIX-S365A (S195A in chymotrypsin numbering) variant, and the hyperactive FIX-R338L variant that exhibits decreased FIX activity with emicizumab, as previously shown with purified protein.14
Summary of clinical and recombinant data of FVIII mimetic–rescuable HB FIX variants
| Variant . | Chymotrypsin no. . | FIX domain . | Recombinant FIX . | Clinical data . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Activity (% WT) . | Antigen (% WT) . | Clot time Enh∗ . | TGA Enh∗ . | Activity (%) . | Antigen (%) . | Reported patients . | |||
| N2I | NA | Gla | 3 | 100 | 3 | 4 | 1 | 60 | 1 |
| K5E | NA | Gla | 5 | 190 | 3 | 4 | 4-7 | NR | 2 |
| L6S | NA | Gla | 2 | 140 | 3 | 6 | <1 | NR | 3 |
| F25C | NA | Gla | 6 | 50 | 2 | 2 | 10 | NR | 1 |
| F25S | NA | Gla | 13 | 150 | 3 | 8 | 2-5 | 25-32 | 5 |
| D47E | NA | EGF1 | 9 | 100 | 7 | 4 | 1-14 | 80-90 | 2 |
| D47G | NA | EGF1 | 8 | 30 | 4 | 1 | 10-24 | 100 | 2 |
| N67K | NA | EGF1 | 35 | 60 | 2 | 1 | <1-12 | NR | 2 |
| E78K | NA | EGF1 | 2 | 40 | 2 | 2 | <1 | <1-84 | 5 |
| E83K | NA | EGF1 | <1 | 10 | 11 | 2 | <1 | <10 | 1 |
| N92K | NA | EGF2 | <1 | 30 | 30 | 18 | <1-2 | 86 | 4 |
| N92S | NA | EGF2 | 3 | 50 | 11 | 25 | NR | NR | 2 |
| V182A | 17 | Protease | 6 | 70 | 3 | 1 | 23 | 100 | 1 |
| V182F | 17 | Protease | <1 | 100 | 3 | 2 | <1 | 120 | 2 |
| V182L | 17 | Protease | 2 | 30 | 9 | 3 | 4-15 | 132 | 3 |
| D269E | 102 | Protease | <1 | 5 | 3 | 2 | <1 | 5 | 1 |
| Y295C | 128 | Protease | <1 | 10 | 4 | 4 | <1 | NR | 2 |
| D332Y | 164 | Protease | 3 | 20 | 4 | 1 | 3-4 | NR | 3 |
| R333L | 165 | Protease | <1 | 30 | 87 | 17 | 2 | 150 | 2 |
| R333P | 165 | Protease | 1 | 40 | 49 | 6 | 1.7 | NR | 1 |
| R333Q | 165 | Protease | 1 | 110 | 49 | 8 | <1-16 | 32-148 | 69 |
| R338P | 170 | Protease | 7 | 80 | 6 | 3 | 10-16 | 27 | 3 |
| T340I | 172 | Protease | 1 | 30 | 17 | 4 | NR | NR | 2 |
| T340R | 172 | Protease | <1 | 80 | 9 | 6 | <1 | NR | 1 |
| K341E | 173 | Protease | 5 | 100 | 26 | 2 | <1-12 | NR | 8 |
| K341N | 173 | Protease | <1 | 80 | 64 | 11 | <1 | 55 | 1 |
| I344F | 176 | Protease | 1 | 70 | 13 | 2 | <1-4 | 87 | 4 |
| I344N | 176 | Protease | <1 | 60 | 22 | 13 | NR | NR | 1 |
| I344S | 176 | Protease | <1 | 60 | 13 | 15 | <1-4 | 112-119 | 2 |
| I344T | 176 | Protease | <1 | 60 | 13 | 5 | 1-2 | 28 | 2 |
| M348I | 180 | Protease | <1 | 80 | 4 | 11 | <1 | 40-93 | 4 |
| M348K | 180 | Protease | <1 | 40 | 3 | 1 | <1-2 | 12 | 2 |
| M348R | 180 | Protease | <1 | 50 | 14 | 3 | NR | NR | 1 |
| G356R | 187 | Protease | 1 | 160 | 24 | 2 | <1-4 | 75 | 9 |
| P368H | 198 | Protease | <1 | 50 | 7 | 6 | <1 | 102 | 1 |
| P368R | 198 | Protease | 10 | 60 | 8 | 1 | <1 | NR | 1 |
| P368T | 198 | Protease | 1 | 130 | 7 | 7 | <1-7 | 100-156 | 13 |
| E387A | 217 | Protease | 2 | 170 | 7 | 2 | <1-4 | 90-118 | 6 |
| E387G | 217 | Protease | 2 | 390 | 24 | 16 | 2 | 95 | 2 |
| E387K | 217 | Protease | 1 | 60 | 17 | 4 | <1-2 | 63-74 | 10 |
| I397T | 227 | Protease | 2 | 70 | 6 | 6 | <1-12 | 27-160 | 44 |
| Variant . | Chymotrypsin no. . | FIX domain . | Recombinant FIX . | Clinical data . | |||||
|---|---|---|---|---|---|---|---|---|---|
| Activity (% WT) . | Antigen (% WT) . | Clot time Enh∗ . | TGA Enh∗ . | Activity (%) . | Antigen (%) . | Reported patients . | |||
| N2I | NA | Gla | 3 | 100 | 3 | 4 | 1 | 60 | 1 |
| K5E | NA | Gla | 5 | 190 | 3 | 4 | 4-7 | NR | 2 |
| L6S | NA | Gla | 2 | 140 | 3 | 6 | <1 | NR | 3 |
| F25C | NA | Gla | 6 | 50 | 2 | 2 | 10 | NR | 1 |
| F25S | NA | Gla | 13 | 150 | 3 | 8 | 2-5 | 25-32 | 5 |
| D47E | NA | EGF1 | 9 | 100 | 7 | 4 | 1-14 | 80-90 | 2 |
| D47G | NA | EGF1 | 8 | 30 | 4 | 1 | 10-24 | 100 | 2 |
| N67K | NA | EGF1 | 35 | 60 | 2 | 1 | <1-12 | NR | 2 |
| E78K | NA | EGF1 | 2 | 40 | 2 | 2 | <1 | <1-84 | 5 |
| E83K | NA | EGF1 | <1 | 10 | 11 | 2 | <1 | <10 | 1 |
| N92K | NA | EGF2 | <1 | 30 | 30 | 18 | <1-2 | 86 | 4 |
| N92S | NA | EGF2 | 3 | 50 | 11 | 25 | NR | NR | 2 |
| V182A | 17 | Protease | 6 | 70 | 3 | 1 | 23 | 100 | 1 |
| V182F | 17 | Protease | <1 | 100 | 3 | 2 | <1 | 120 | 2 |
| V182L | 17 | Protease | 2 | 30 | 9 | 3 | 4-15 | 132 | 3 |
| D269E | 102 | Protease | <1 | 5 | 3 | 2 | <1 | 5 | 1 |
| Y295C | 128 | Protease | <1 | 10 | 4 | 4 | <1 | NR | 2 |
| D332Y | 164 | Protease | 3 | 20 | 4 | 1 | 3-4 | NR | 3 |
| R333L | 165 | Protease | <1 | 30 | 87 | 17 | 2 | 150 | 2 |
| R333P | 165 | Protease | 1 | 40 | 49 | 6 | 1.7 | NR | 1 |
| R333Q | 165 | Protease | 1 | 110 | 49 | 8 | <1-16 | 32-148 | 69 |
| R338P | 170 | Protease | 7 | 80 | 6 | 3 | 10-16 | 27 | 3 |
| T340I | 172 | Protease | 1 | 30 | 17 | 4 | NR | NR | 2 |
| T340R | 172 | Protease | <1 | 80 | 9 | 6 | <1 | NR | 1 |
| K341E | 173 | Protease | 5 | 100 | 26 | 2 | <1-12 | NR | 8 |
| K341N | 173 | Protease | <1 | 80 | 64 | 11 | <1 | 55 | 1 |
| I344F | 176 | Protease | 1 | 70 | 13 | 2 | <1-4 | 87 | 4 |
| I344N | 176 | Protease | <1 | 60 | 22 | 13 | NR | NR | 1 |
| I344S | 176 | Protease | <1 | 60 | 13 | 15 | <1-4 | 112-119 | 2 |
| I344T | 176 | Protease | <1 | 60 | 13 | 5 | 1-2 | 28 | 2 |
| M348I | 180 | Protease | <1 | 80 | 4 | 11 | <1 | 40-93 | 4 |
| M348K | 180 | Protease | <1 | 40 | 3 | 1 | <1-2 | 12 | 2 |
| M348R | 180 | Protease | <1 | 50 | 14 | 3 | NR | NR | 1 |
| G356R | 187 | Protease | 1 | 160 | 24 | 2 | <1-4 | 75 | 9 |
| P368H | 198 | Protease | <1 | 50 | 7 | 6 | <1 | 102 | 1 |
| P368R | 198 | Protease | 10 | 60 | 8 | 1 | <1 | NR | 1 |
| P368T | 198 | Protease | 1 | 130 | 7 | 7 | <1-7 | 100-156 | 13 |
| E387A | 217 | Protease | 2 | 170 | 7 | 2 | <1-4 | 90-118 | 6 |
| E387G | 217 | Protease | 2 | 390 | 24 | 16 | 2 | 95 | 2 |
| E387K | 217 | Protease | 1 | 60 | 17 | 4 | <1-2 | 63-74 | 10 |
| I397T | 227 | Protease | 2 | 70 | 6 | 6 | <1-12 | 27-160 | 44 |
Clinical data extracted from FIX-variant database.15
NR, not reported.
Clot time and TGA enhancement (Enh) are the ratios of the parameter with and without emicizumab as detailed in article text.
The FVIII-mimetic enhancement is independent of the amount of FIX present in the media, because there was no relationship between the antigen level and activity enhancement (Figure 1C; Spearman coefficient −0.03; P = .9). Likewise, the increase in FIX activity (Figure 1A) is specific to emicizumab because supratherapeutic levels of FVIII (100% normal) failed to enhance FIX activity (Figure 1D).
We also tested the ability of emicizumab to improve thrombin production in these FIX variants (Figure 1E; supplemental Figures 3-5). The TGA has been used to demonstrate that emicizumab has a procoagulant effect in preclinical and clinical HA studies22,23 and to safely customize FVIII-mimetic treatment.24 We found that emicizumab enhanced thrombin generation in most of these LOF FIX variants by up to 30-fold (Table 1). There is a strong and moderate correlation between the FIX activity and the peak thrombin level in the rescuable FIX variants with and without emicizumab, respectively (supplemental Figure 6). Biochemical characterization of the most common rescuable variant (FIX-R333Q) with purified recombinant FIX protein demonstrated similar and dose-dependent procoagulant enhancement with emicizumab and an impaired interaction between FIXa and FVIIIa (supplemental Figure 7) similar to previous studies.20
We also evaluated the ability of emicizumab to improve ex vivo clotting in samples from patients with HB. Patient 1 had moderate HB (FIX activity 1%-3% of normal) owing to a FIX-I397T variant and an annualized bleeding rate of 1.7 bleeds per year, consistent with the natural history of moderate HB.25 His FIX activity and antigen level were 3% and 100%, respectively. His activated partial thromboplastin time-based clotting time decreased with increasing amounts of emicizumab, whereas the clotting time of control HB plasma with undetectable FIX antigen did not change (Figure 1F). Patient 2 also had a FIX-I397T variant and 3% FIX activity. Rotational thromboelastometry measurements of whole blood from patient 2 demonstrated improved clotting time with the addition of emicizumab, namely from 1229–1330 seconds to 607–888 seconds (95% confidence interval) (Figure 1G). This improvement in both plasma and whole blood clot formation from clinical samples with a FIX-I397T variant supports the translational relevance of the screen in Figure 1A.
Combined, these data demonstrate the potential of emicizumab and likely other FVIII mimetics to improve the procoagulant activity of selected HB-causing FIX variants. Importantly, this improvement cannot exceed the activity of FIX-WT. The identified FIX variants have amino acid substitutions at positions within the FVIIIa-binding site (supplemental Figure 1A) or at positions implicated in allostery (supplemental Figure 1B); as expected, not every variant with substitutions within these regions responded to emicizumab, because substitutions may disrupt enzyme function through several mechanisms. Substitutions outside of these motifs, such as in the catalytic site (S365A) or the protease calcium–binding site, do not have improved procoagulant activity with emicizumab (Figure 1B). The magnitude of some of these improvements may be translationally relevant. As such, FVIII mimetics may represent an alternative NFT for HB with these select F9 genotypes.
Acknowledgments
The authors thank Leslie Raffini for assistance in recruitment of patients for the study and Sebastian E. Leyes Porello for work purifying recombinant FIX variants. They acknowledge helpful discussion and/or editorial comments from Valder Arruda, Rodney Camire, and Sriram Krishnaswamy.
B.J.S.-J. received support from the National Blood Foundation and National Institutes of Health, National Heart, Lung, and Blood Institute (grant K08HL14007). This study is also supported by the Frontiers Program at the Children’s Hospital of Philadelphia.
Authorship
Contribution: K.L., J.Q.C., Y.B.S., and A.R.S. designed and conducted the experiments; A.M.P. provided clinical histories, samples, and guidance, and revised the manuscript; L.A.G. designed the experiments, interpreted the data, provided guidance, and revised the manuscript; V.G.B. and B.S.D. supervised clinical recruitment, provided guidance, and revised the manuscript; and B.J.S.-J. designed and conducted the experiments, analyzed and interpreted the data, supervised the project, and drafted the manuscript.
Conflict-of-interest disclosure: B.J.S.-J. is on an advisory board sponsored by Genentech, unrelated to this work. The remaining authors declare no competing financial interests.
Correspondence: Benjamin J. Samelson-Jones, Children’s Hospital of Philadelphia, Colket Translational Research Center, Colket Translational Research Center Room 5028, 3501 Civic Center Blvd, Philadelphia, PA 19104; email: samelsonjonesb@chop.edu.
References
Author notes
K.L. and J.Q.C. are joint first authors.
Original data are available on request from the corresponding author, Benjamin J. Samelson-Jones (samelsonjonesb@chop.edu).
The online version of this article contains a data supplement.

![Evaluation of procoagulant enhancement of HB-causing FIX variants by emicizumab. (A) The FIX activity of conditioned media from HEK293 cells that transiently expressed recombinant select HB-causing FIX variants with 0 (gray) or 300 nM (red) emicizumab. Only variants with a greater than or equal to twofold enhancement with emicizumab are shown; nonrescuable variants are listed in supplemental Table 1. For clarity, variants with FIX activity with emicizumab >5% of normal are shown on the left, whereas variants with FIX activity with emicizumab 1% to 5% of normal are shown on the right. FIX activity was determined using an activated partial thromboplastin time (aPTT)–based assay with standard curves for 0 or 300 nM emicizumab (supplemental Figure 2). Values are normalized to recombinant FIX-WT activity expressed in the same experiment. Bars represent the mean ± standard error of mean of ≥9 independent transfections. (B) FIX activity of conditioned media with recombinant FIX protein controls with 0 (gray) or 300 nM emicizumab (blue). (C) Enhancement with emicizumab as a function of recombinant FIX antigen. (D) Enhancement with 300 nM emicizumab (red) or 1000% normal FVIII (light blue) of select HB-causing FIX variants. (E) Thrombograms of FIX variants with 0 nM (gray) or 300 nM emicizumab (red). Thrombograms of controls FIX-WT and UC with 0 nM (gray) or 300 nM emicizumab (blue). Coagulation was triggered with FXIa. Results are representative of ≥2 thrombograms from ≥2 independent transfections. Additional thrombograms of rescuable HB-causing variants are provided in supplemental Figure 3. (F) An aPTT-based clotting time of plasma from patients with moderate HB with FIX-I397T variant (patient 1) with increasing amounts of emicizumab in comparison with commercial severe HB plasma with undetectable FIX antigen (cross reactive material [CRM] negative). Points indicate the mean ± standard deviation of 4 measurements. The horizontal gray line represents the clotting time of normal human plasma. (G) Rotational thromboelastometry (ROTEM) thromboelastograms of whole blood from a patient with moderate HB with an FIX-I397T variant (patient 2) with 300 nM emicizumab or 100% recombinant FIX added ex vivo. Results are representative of 2 measurements. UC, unconditioned media.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/11/10.1182_blood.2023021944/2/m_blood_bld-2023-021944-gr1fg.jpeg?Expires=1767733847&Signature=q5~BPPaVvkSBrQ6Aq51p80ZUYLzpqxqD5JkOxllOHlp7cnelgrhEDbVBKrMNgGQtCJ~ZJqbvRYEbCQA91PG1CYCffQ65GrUHlyogXRL0NP0OicSuMp0gVggi~4n4GGJu8NKQFdb36DLzYysrV6Jq0ko37~1LcmtxXbw~H~xZ8glnWcjlPkc8gD6IYmnuyGYSVZJFNPcYhdYPJoO1Z0l2ZMfRfV5XlKmaiSiZQyILZs3AuhZL1uR6kFZml0UXwiKt8opt0RcuxdJPr76D2JzX4joZ3zj-DX7Ka~Gc-SGZHp6B3U7dzG-NmlbSOLcM33ZN86iaFmuiUQicwNS7tym85w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal