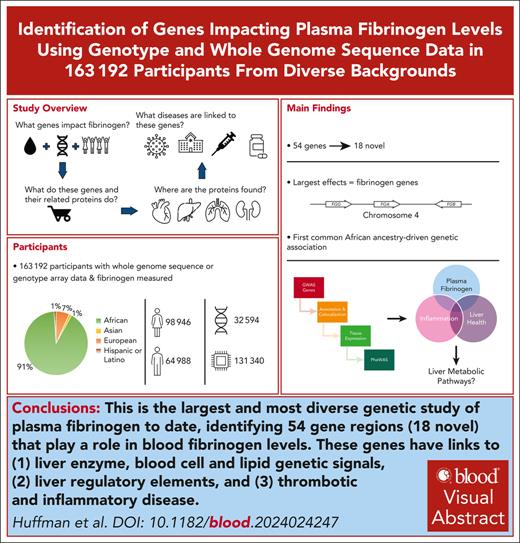
Key Points
Largest and most diverse genetic study of plasma fibrinogen identifies 54 regions (18 novel), housing 69 distinct variants (20 novel).
Links to (1) liver enzyme, blood cell, and lipid genetic signals, (2) liver regulatory elements, and (3) thrombotic and inflammatory disease.
Visual Abstract
Genetic studies have identified numerous regions associated with plasma fibrinogen levels in Europeans, yet missing heritability and limited inclusion of non-Europeans necessitates further studies with improved power and sensitivity. Compared with array-based genotyping, whole-genome sequencing (WGS) data provide better coverage of the genome and better representation of non-European variants. To better understand the genetic landscape regulating plasma fibrinogen levels, we meta-analyzed WGS data from the National Heart, Lung, and Blood Institute’s Trans-Omics for Precision Medicine (TOPMed) program (n = 32 572), with array-based genotype data from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (n = 131 340) imputed to the TOPMed or Haplotype Reference Consortium panel. We identified 18 loci that have not been identified in prior genetic studies of fibrinogen. Of these, 4 are driven by common variants of small effect with reported minor allele frequency (MAF) at least 10 percentage points higher in African populations. Three signals (SERPINA1, ZFP36L2, and TLR10) contain predicted deleterious missense variants. Two loci, SOCS3 and HPN, each harbor 2 conditionally distinct, noncoding variants. The gene region encoding the fibrinogen protein chain subunits (FGG;FGB;FGA) contains 7 distinct signals, including 1 novel signal driven by rs28577061, a variant common in African ancestry populations but extremely rare in Europeans (MAFAFR = 0.180; MAFEUR = 0.008). Through phenome-wide association studies in the VA Million Veteran Program, we found associations between fibrinogen polygenic risk scores and thrombotic and inflammatory disease phenotypes, including an association with gout. Our findings demonstrate the utility of WGS to augment genetic discovery in diverse populations and offer new insights for putative mechanisms of fibrinogen regulation.
Introduction
Fibrinogen is a critical coagulation factor and acute phase reactive protein. Under normal conditions, fibrinogen is abundant in circulation, yet during the acute phase inflammatory response, interleukin--6 (IL-6) and IL-1 mediate transcriptional cascades that increase circulating fibrinogen levels up to 3-fold above baseline.1,2 Fibrinogen measures are a clinical predictor of thrombotic diseases, including coronary heart disease, myocardial infarction, venous thromboembolism, and ischemic stroke.3,4 Although animal models have shown a causative relationship between fibrinogen and thrombosis,5,6 this has been difficult to confirm in human genetic studies.7-9
Circulating fibrinogen levels are estimated to be 21% to 67% heritable,10,11 with most estimates falling in the 30% to 50% range,12,13 and heterogeneous across diverse populations.14-16 Individuals identifying as African American have higher reported baseline levels of fibrinogen,14-16 with 1 study suggesting higher fibrinogen heritability in African Americans (44%) vs non-Hispanic Whites (28%).17 Although genome-wide and exome-wide sequencing studies have identified several loci associated with fibrinogen measures, these variants explain a maximum of 3.7% of variance in European populations.7,18,19 Little is known regarding genetic regulation of fibrinogen across diverse populations.
Unlike genotyping arrays, which often have better coverage of variants common in European populations, whole-genome sequencing (WGS) allows nontargeted genomic interrogation across all populations.20 Broader coverage provided by WGS increases confidence in minor allele calls, improving power to detect associations with rare and low-frequency variants, and to distinguish multiple signals in the same region.21,22 Furthermore, large-scale WGS-based reference panels improve imputation quality, increasing power in meta-analysis.21,22 Incorporating more sensitive genomic measurements, such as WGS, in genetic analyses of fibrinogen may reveal additional associations across a range of allele frequencies and effect sizes in diverse populations.
In this study, we performed genome-wide analyses integrating WGS data from National Heart, Lung, and Blood Institute’s Trans-Omics for Precision Medicine (TOPMed) program23 and TOPMed-imputed genotyping data from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium24 to identify new genetic variants associated with circulating fibrinogen. To determine putative regulatory mechanisms and prioritize potentially causal genes, we performed in silico annotation, colocalization analyses, transcriptome-wide association study (TWAS), and TWAS fine mapping. Additionally, we tested the hypothesis that polygenic risk scores (PRSs) for fibrinogen are also associated with coagulation and inflammation-related phenotypes in European and African ancestry participants from the VA Million Veteran Program (MVP).25
Materials and methods
Design and study population
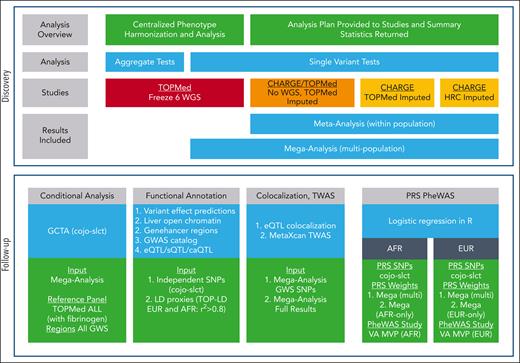
To investigate the genetic architecture of circulating fibrinogen, we performed a multipopulation genome-wide association study (GWAS), followed by TWAS and phenome-wide association study (PheWAS). An overview of the study design is in Figure 1 and supplemental Figure 1, available on the Blood website. These analyses were undertaken within National Heart, Lung, and Blood Institute’s TOPMed Program and the CHARGE Consortium’s Hemostasis Working Group. In total, 163 912 participants contributed to primary genomic analyses, including 11 283 African-ancestry (AFR), 741 Asian-ancestry, 149 619 European-ancestry (EUR), and 2061 Hispanic participants. Details of the 40 participating studies are in Table 1 and the supplemental Data. The contributing studies were all approved by their relevant institutional review boards.
Participating studies
| Source . | Category . | Study . | Group . | Population group . | Total no. . | % Male . | Age, mean (SD), y . | Fibrinogen mean (SD), g/L . | Fibrinogen method . |
|---|---|---|---|---|---|---|---|---|---|
| TOPMed | Freeze 6 WGS | Amish Research Program | EUR | 558 | 48.0 | 56.3 (14.2) | 3.2 (0.8) | Clauss | |
| Atherosclerosis Risk in Communities (ARIC) | AFR | 664 | 40.5 | 54.2 (6.0) | 3.3 (0.8) | Clauss | |||
| EUR | 5 963 | 45.7 | 54.0 (5.7) | 3.0 (0.6) | |||||
| Coronary Artery Risk Development in Young Adults Study (CARDIA) | AFR | 542 | 39.5 | 26.9 (4.2) | 2.3 (0.5) | Clauss | |||
| EUR | 874 | 45.7 | 28.3 (3.4) | 2.1 (0.5) | |||||
| NA | 131 | 52.7 | 27.0 (3.9) | 2.3 (0.6) | |||||
| Cleveland Family Study (CFS) | AFR | 366 | 43.4 | 40.2 (18.4) | 3.2 (0.9) | Clauss | |||
| EUR | 258 | 47.7 | 45.9 (19.1) | 3.3 (0.7) | |||||
| Cardiovascular Health Study (CHS) | AFR | 497 | 39.4 | 72.4 (5.2) | 3.4 (0.7) | Clauss | |||
| EUR | 2 247 | 44.4 | 72.5 (5.3) | 3.2 (0.7) | |||||
| COPDGene Study | AFR | 188 | 51.6 | 54.3 (7.0) | 5.0 (1.1) | Clauss | |||
| EUR | 1 191 | 48.7 | 62.0 (9.1) | 5.1 (1.2) | |||||
| Framingham Heart Study (FHS) | EUR | 3 535 | 46.2 | 49.2 (11.9) | 3.2 (0.6) | Clauss | |||
| Genetic Study of Atherosclerosis Risk (GeneSTAR) | AFR | 767 | 36.4 | 41.0 (10.9) | 3.8 (1.1) | Clauss | |||
| EUR | 945 | 44.2 | 42.7 (12.3) | 3.6 (1.1) | |||||
| Genetic Epidemiology Network of Arteriopathy (GENOA) | AFR | 703 | 26.6 | 61.0 (10.4) | 3.8 (1.0) | Clauss | |||
| Jackson Heart Study (JHS) | AFR | 2 926 | 38.0 | 54.9 (12.9) | 4.2 (0.9) | Clauss | |||
| Multi-Ethnic Study of Atherosclerosis (MESA) | AFR | 1 082 | 47.2 | 60.8 (9.6) | 3.6 (0.8) | Clauss | |||
| ASN | 1 828 | 49.1 | 61.5 (9.8) | 3.3 (0.7) | |||||
| EUR | 1 014 | 48.6 | 60.2 (9.8) | 3.6 (0.7) | |||||
| HIS | 601 | 50.2 | 61.2 (10.1) | 3.3 (0.6) | |||||
| San Antonio Family Study (SAFS) | HIS | 370 | 35.9 | 41.0 (15.8) | 3.0 (0.7) | Clauss | |||
| NA | 22 | 45.5 | 43.0 (21.6) | 3.2 (1.0) | |||||
| Women’s Health Initiative (WHI) | AFR | 740 | 0.0 | 63.0 (7.0) | 3.2 (0.7) | Clauss | |||
| AI/AN | 35 | 0.0 | 64.0 (7.7) | 3.1 (0.7) | |||||
| ASN | 140 | 0.0 | 66.2 (7.4) | 2.9 (0.6) | |||||
| EUR | 4 213 | 0.0 | 67.2 (6.9) | 3.0 (0.7) | |||||
| HIS | 174 | 0.0 | 62.5 (6.2) | 3.1 (0.7) | |||||
| NA | 20 | 0.0 | 68.0 (7.3) | 3.2 (0.9) | |||||
| No WGS, TOPMed Imputation | ARIC | AFR | 2 170 | 36.4 | 53.2 (5.7) | 3.2 (0.7) | See above | ||
| EUR | 3 790 | 48.6 | 54.6 (5.7) | 3.0 (0.6) | |||||
| CHS | EUR | 1 508 | 38.9 | 72.3 (5.4) | 3.2 (0.6) | See above | |||
| FHS | EUR | 1 565 | 44.5 | 49.5 (11.9) | 3.2 (0.7) | See above | |||
| WHI | GARNET | EUR | 442 | 0.0 | 64.7 (7.5) | 3.2 (0.9) | See above | ||
| ONCO | EUR | 312 | 0.0 | 66.9 (5.8) | 3.1 (0.9) | ||||
| HIPFX | EUR | 133 | 0.0 | 70.5 (4.9) | 3.3 (1.0) | ||||
| SHARE | AFR | 496 | 0.0 | 62.1 (7.3) | 3.5 (0.9) | ||||
| HIS | 357 | 0.0 | 60.1 (6.8) | 3.2 (0.7) | |||||
| CHARGE | TOPMed Imputation | Airwave Health Monitoring Study (Airwave) | EUR | 13 405 | 65.8 | 40.4 (9.0) | 3.8 (1.0) | Clauss | |
| Caerphilly Prospective study (CaPs) | EUR | 1 159 | 100.0 | 56.8 (4.5) | 4.0 (0.8) | Clauss and HN | |||
| CHRIS | EUR | 9 012 | 43.9 | 44.2 (15.4) | 2.8 (0.6) | Clauss | |||
| CROATIA | Korcula | EUR | 798 | 35.4 | 56.3 (14.0) | 4.6 (1.5) | Clauss | ||
| Split | EUR | 953 | 38.9 | 50.3 (14.0) | 4.1 (1.6) | Clauss | |||
| Vis | EUR | 924 | 42.5 | 56.1 (15.6) | 3.6 (0.8) | Clauss | |||
| European Prospective Investigation in Cancer (EPIC) Norfolk Study (EPIC-Norfolk) | EUR | 17 473 | 47.0 | 59.2 (9.2) | 2.9 (1.0) | Clauss | |||
| FHS_omni | AFR | 142 | 37.3 | 53.5 (9.4) | 3.4 (0.7) | Clauss | |||
| HIS | 168 | 36.3 | 49.1 (8.0) | 3.3 (0.6) | |||||
| Genetic Analysis of Idiopathic Thrombophilia (GAIT2) | EUR | 896 | 49.4 | 39.7 (21.4) | 3.5 (0.8) | Clauss | |||
| Helsinki Birth Cohort Study (HBCS) | EUR | 1 321 | 40.6 | 61.4 (2.9) | 3.3 (1.0) | Clauss | |||
| Lothian Birth Cohort 1921 (LBC1921) | EUR | 486 | 42.4 | 76.1 (0.6) | 3.6 (0.9) | Clauss | |||
| Lothian Birth Cohort 1936 (LBC1936) | EUR | 988 | 50.8 | 69.6 (0.8) | 3.3 (0.6) | Clauss | |||
| Ludwigshafen Risk and Cardiovascular Health (LURIC) | EUR | 2 731 | 70.0 | 62.7 (10.6) | 4.0 (1.1) | Clauss | |||
| Marseille Thrombosis Association (MARTHA) | EUR | 985 | 29.0 | 45.4 (14.9) | 3.4 (0.7) | Clauss | |||
| Netherlands Epidemiology of Obesity (NEO) | EUR | 5 303 | 46.8 | 56.0 (6.0) | 3.0 (0.6) | Clauss | |||
| Netherlands Twin Registry (NTR) | EUR | 7 650 | 38.5 | 44.9 (14.6) | 2.8 (0.7) | Clauss | |||
| Precocious Coronary Artery Disease Study (PROCARDIS) | Cases | EUR | 4 196 | 73.0 | 62.2 (7.0) | 4.2 (1.0) | Clauss and IN | ||
| Controls | EUR | 2 224 | 76.2 | 55.1 (8.3) | 3.5 (0.9) | ||||
| Prospective Study of Pravastatin in the Elderly at Risk (PROSPER) | EUR | 4 951 | 48.1 | 75.3 (3.3) | 3.6 (0.7) | Clauss | |||
| RETROVE | Cases | EUR | 398 | 49.2 | 61.8 (18.6) | 3.8 (0.9) | Clauss | ||
| Controls | EUR | 398 | 48.7 | 47.1 (18.2) | 3.4 (0.7) | ||||
| SardiNIA | EUR | 5 931 | 35.9 | 43.6 (17.6) | 3.3 (0.7) | Clauss | |||
| Study of Health in Pomerania (SHIP) | SHIP-START | EUR | 3 571 | 47.5 | 47.5 (15.6) | 3.0 (0.7) | Clauss | ||
| SHIP-TREND-B1 | EUR | 909 | 42.9 | 49.1 (13.4) | 3.0 (0.7) | Clauss | |||
| SHIP-TREND-B2 | EUR | 1 505 | 49.0 | 49.5 (15.0) | 3.0 (0.8) | Clauss | |||
| TWINSUK | EUR | 2 037 | 4.7 | 49.1 (12.6) | 3.0 (0.8) | Clauss | |||
| Viking Health Study-Shetland (VIKING) | EUR | 2 086 | 40.1 | 49.9 (15.2) | 3.5 (0.8) | Clauss | |||
| Women’s Genome Health Study (WGHS) | EUR | 22 962 | 0.0 | 54.2 (7.1) | 3.6 (0.8) | ITA | |||
| HRC Imputation | GOYA | EUR | 1 431 | 100.0 | 45.6 (7.9) | 3.1 (0.8) | FPA | ||
| Inter99 | EUR | 649 | 61.8 | 49.5 (6.4) | 2.1 (1.5) | UI | |||
| Rotterdam Study (RS) | RS1 | EUR | 2 252 | 36.5 | 70.5 (8.9) | 2.8 (0.7) | Clauss | ||
| RS2 | EUR | 673 | 45.5 | 64.8 (8.0) | 3.9 (0.9) | ||||
| Source . | Category . | Study . | Group . | Population group . | Total no. . | % Male . | Age, mean (SD), y . | Fibrinogen mean (SD), g/L . | Fibrinogen method . |
|---|---|---|---|---|---|---|---|---|---|
| TOPMed | Freeze 6 WGS | Amish Research Program | EUR | 558 | 48.0 | 56.3 (14.2) | 3.2 (0.8) | Clauss | |
| Atherosclerosis Risk in Communities (ARIC) | AFR | 664 | 40.5 | 54.2 (6.0) | 3.3 (0.8) | Clauss | |||
| EUR | 5 963 | 45.7 | 54.0 (5.7) | 3.0 (0.6) | |||||
| Coronary Artery Risk Development in Young Adults Study (CARDIA) | AFR | 542 | 39.5 | 26.9 (4.2) | 2.3 (0.5) | Clauss | |||
| EUR | 874 | 45.7 | 28.3 (3.4) | 2.1 (0.5) | |||||
| NA | 131 | 52.7 | 27.0 (3.9) | 2.3 (0.6) | |||||
| Cleveland Family Study (CFS) | AFR | 366 | 43.4 | 40.2 (18.4) | 3.2 (0.9) | Clauss | |||
| EUR | 258 | 47.7 | 45.9 (19.1) | 3.3 (0.7) | |||||
| Cardiovascular Health Study (CHS) | AFR | 497 | 39.4 | 72.4 (5.2) | 3.4 (0.7) | Clauss | |||
| EUR | 2 247 | 44.4 | 72.5 (5.3) | 3.2 (0.7) | |||||
| COPDGene Study | AFR | 188 | 51.6 | 54.3 (7.0) | 5.0 (1.1) | Clauss | |||
| EUR | 1 191 | 48.7 | 62.0 (9.1) | 5.1 (1.2) | |||||
| Framingham Heart Study (FHS) | EUR | 3 535 | 46.2 | 49.2 (11.9) | 3.2 (0.6) | Clauss | |||
| Genetic Study of Atherosclerosis Risk (GeneSTAR) | AFR | 767 | 36.4 | 41.0 (10.9) | 3.8 (1.1) | Clauss | |||
| EUR | 945 | 44.2 | 42.7 (12.3) | 3.6 (1.1) | |||||
| Genetic Epidemiology Network of Arteriopathy (GENOA) | AFR | 703 | 26.6 | 61.0 (10.4) | 3.8 (1.0) | Clauss | |||
| Jackson Heart Study (JHS) | AFR | 2 926 | 38.0 | 54.9 (12.9) | 4.2 (0.9) | Clauss | |||
| Multi-Ethnic Study of Atherosclerosis (MESA) | AFR | 1 082 | 47.2 | 60.8 (9.6) | 3.6 (0.8) | Clauss | |||
| ASN | 1 828 | 49.1 | 61.5 (9.8) | 3.3 (0.7) | |||||
| EUR | 1 014 | 48.6 | 60.2 (9.8) | 3.6 (0.7) | |||||
| HIS | 601 | 50.2 | 61.2 (10.1) | 3.3 (0.6) | |||||
| San Antonio Family Study (SAFS) | HIS | 370 | 35.9 | 41.0 (15.8) | 3.0 (0.7) | Clauss | |||
| NA | 22 | 45.5 | 43.0 (21.6) | 3.2 (1.0) | |||||
| Women’s Health Initiative (WHI) | AFR | 740 | 0.0 | 63.0 (7.0) | 3.2 (0.7) | Clauss | |||
| AI/AN | 35 | 0.0 | 64.0 (7.7) | 3.1 (0.7) | |||||
| ASN | 140 | 0.0 | 66.2 (7.4) | 2.9 (0.6) | |||||
| EUR | 4 213 | 0.0 | 67.2 (6.9) | 3.0 (0.7) | |||||
| HIS | 174 | 0.0 | 62.5 (6.2) | 3.1 (0.7) | |||||
| NA | 20 | 0.0 | 68.0 (7.3) | 3.2 (0.9) | |||||
| No WGS, TOPMed Imputation | ARIC | AFR | 2 170 | 36.4 | 53.2 (5.7) | 3.2 (0.7) | See above | ||
| EUR | 3 790 | 48.6 | 54.6 (5.7) | 3.0 (0.6) | |||||
| CHS | EUR | 1 508 | 38.9 | 72.3 (5.4) | 3.2 (0.6) | See above | |||
| FHS | EUR | 1 565 | 44.5 | 49.5 (11.9) | 3.2 (0.7) | See above | |||
| WHI | GARNET | EUR | 442 | 0.0 | 64.7 (7.5) | 3.2 (0.9) | See above | ||
| ONCO | EUR | 312 | 0.0 | 66.9 (5.8) | 3.1 (0.9) | ||||
| HIPFX | EUR | 133 | 0.0 | 70.5 (4.9) | 3.3 (1.0) | ||||
| SHARE | AFR | 496 | 0.0 | 62.1 (7.3) | 3.5 (0.9) | ||||
| HIS | 357 | 0.0 | 60.1 (6.8) | 3.2 (0.7) | |||||
| CHARGE | TOPMed Imputation | Airwave Health Monitoring Study (Airwave) | EUR | 13 405 | 65.8 | 40.4 (9.0) | 3.8 (1.0) | Clauss | |
| Caerphilly Prospective study (CaPs) | EUR | 1 159 | 100.0 | 56.8 (4.5) | 4.0 (0.8) | Clauss and HN | |||
| CHRIS | EUR | 9 012 | 43.9 | 44.2 (15.4) | 2.8 (0.6) | Clauss | |||
| CROATIA | Korcula | EUR | 798 | 35.4 | 56.3 (14.0) | 4.6 (1.5) | Clauss | ||
| Split | EUR | 953 | 38.9 | 50.3 (14.0) | 4.1 (1.6) | Clauss | |||
| Vis | EUR | 924 | 42.5 | 56.1 (15.6) | 3.6 (0.8) | Clauss | |||
| European Prospective Investigation in Cancer (EPIC) Norfolk Study (EPIC-Norfolk) | EUR | 17 473 | 47.0 | 59.2 (9.2) | 2.9 (1.0) | Clauss | |||
| FHS_omni | AFR | 142 | 37.3 | 53.5 (9.4) | 3.4 (0.7) | Clauss | |||
| HIS | 168 | 36.3 | 49.1 (8.0) | 3.3 (0.6) | |||||
| Genetic Analysis of Idiopathic Thrombophilia (GAIT2) | EUR | 896 | 49.4 | 39.7 (21.4) | 3.5 (0.8) | Clauss | |||
| Helsinki Birth Cohort Study (HBCS) | EUR | 1 321 | 40.6 | 61.4 (2.9) | 3.3 (1.0) | Clauss | |||
| Lothian Birth Cohort 1921 (LBC1921) | EUR | 486 | 42.4 | 76.1 (0.6) | 3.6 (0.9) | Clauss | |||
| Lothian Birth Cohort 1936 (LBC1936) | EUR | 988 | 50.8 | 69.6 (0.8) | 3.3 (0.6) | Clauss | |||
| Ludwigshafen Risk and Cardiovascular Health (LURIC) | EUR | 2 731 | 70.0 | 62.7 (10.6) | 4.0 (1.1) | Clauss | |||
| Marseille Thrombosis Association (MARTHA) | EUR | 985 | 29.0 | 45.4 (14.9) | 3.4 (0.7) | Clauss | |||
| Netherlands Epidemiology of Obesity (NEO) | EUR | 5 303 | 46.8 | 56.0 (6.0) | 3.0 (0.6) | Clauss | |||
| Netherlands Twin Registry (NTR) | EUR | 7 650 | 38.5 | 44.9 (14.6) | 2.8 (0.7) | Clauss | |||
| Precocious Coronary Artery Disease Study (PROCARDIS) | Cases | EUR | 4 196 | 73.0 | 62.2 (7.0) | 4.2 (1.0) | Clauss and IN | ||
| Controls | EUR | 2 224 | 76.2 | 55.1 (8.3) | 3.5 (0.9) | ||||
| Prospective Study of Pravastatin in the Elderly at Risk (PROSPER) | EUR | 4 951 | 48.1 | 75.3 (3.3) | 3.6 (0.7) | Clauss | |||
| RETROVE | Cases | EUR | 398 | 49.2 | 61.8 (18.6) | 3.8 (0.9) | Clauss | ||
| Controls | EUR | 398 | 48.7 | 47.1 (18.2) | 3.4 (0.7) | ||||
| SardiNIA | EUR | 5 931 | 35.9 | 43.6 (17.6) | 3.3 (0.7) | Clauss | |||
| Study of Health in Pomerania (SHIP) | SHIP-START | EUR | 3 571 | 47.5 | 47.5 (15.6) | 3.0 (0.7) | Clauss | ||
| SHIP-TREND-B1 | EUR | 909 | 42.9 | 49.1 (13.4) | 3.0 (0.7) | Clauss | |||
| SHIP-TREND-B2 | EUR | 1 505 | 49.0 | 49.5 (15.0) | 3.0 (0.8) | Clauss | |||
| TWINSUK | EUR | 2 037 | 4.7 | 49.1 (12.6) | 3.0 (0.8) | Clauss | |||
| Viking Health Study-Shetland (VIKING) | EUR | 2 086 | 40.1 | 49.9 (15.2) | 3.5 (0.8) | Clauss | |||
| Women’s Genome Health Study (WGHS) | EUR | 22 962 | 0.0 | 54.2 (7.1) | 3.6 (0.8) | ITA | |||
| HRC Imputation | GOYA | EUR | 1 431 | 100.0 | 45.6 (7.9) | 3.1 (0.8) | FPA | ||
| Inter99 | EUR | 649 | 61.8 | 49.5 (6.4) | 2.1 (1.5) | UI | |||
| Rotterdam Study (RS) | RS1 | EUR | 2 252 | 36.5 | 70.5 (8.9) | 2.8 (0.7) | Clauss | ||
| RS2 | EUR | 673 | 45.5 | 64.8 (8.0) | 3.9 (0.9) | ||||
AFR, African American; AI/AN, American Indian or Alaska Native; ASN, East Asian American; EUR, European or European American; FPA, functional photometric assay; HIS, Hispanic or Latino; HN, heat-nephelometry; IN, immune-nephelometric; ITA, immunoturbidimetric assay; NA, missing; UI, ultrasensitive immunoassay; WGS, whole-genome sequencing.
Population group assignment was determined by each study internally. Studies often used some combination of self-reported race or ethnicity data (which in many cases was used for stratification at the genotyping stage, making future pooled analysis challenging) and comparison of ancestry principal components to commonly used reference panels such as 1000G (often with exclusion of any extreme outliers). We acknowledge that this is not concordant with the most up-to-date standards for defining ancestry clusters based on similarity to reference panels26 but do not have access to individual-level data for most participants and are thus reliant on these study assignments.
Phenotyping and harmonization
Fibrinogen was measured in g/L. Most studies measured levels of functional fibrinogen using the Clauss method,27 whereas the remaining 7 studies used a variety of approaches, including nephelometry and enzyme-linked immunosorbent assay, to measure fibrinogen antigen, a measure of overall fibrinogen quantity. Study-specific phenotyping methods can be found in Table 1 and the supplemental Data. Measures of plasma fibrinogen for TOPMed studies were harmonized to ensure common units (g/L) and absence of unexpected distributions or an excess of outliers (supplemental Figure 1). Data were then uploaded to Analysis Commons28 for centralized genetic analysis.
WGS of TOPMed participants
TOPMed WGS methods were described previously.23 Briefly, WGS was conducted at 6 sequencing centers (mean depth >30×, Illumina HiSeq X Ten instruments). Joint variant discovery and genotype calling were conducted by the TOPMed Informatics Research Center across all TOPMed studies using the GotCloud pipeline, resulting in a single genotype call set encompassing all TOPMed (TOPMed Freeze 6). Variant quality control was also performed centrally by the Informatics Research Center. Sample quality control was performed by the TOPMed Data Coordinating Center. Further details are in the supplemental Data.
Genotype imputation of non-TOPMed studies
Genome-wide association analyses and meta-analyses
TOPMed WGS genetic analyses were conducted using mixed models and inverse normalized and rescaled residuals, adjusting for age, sex, population group × study, TOPMed sequencing phase, study-specific parameters, and 10 ancestry-informative principal components. Single variant and aggregate gene-based tests were implemented using the SMMAT function of GENESIS on the Analysis Commons cloud computing platform.28,30 Aggregate tests included only variants with minor allele frequency (MAF) <5% and minor allele count (MAC) ≥1 and used 3 strategies for variant selection: (1) loss of function (LOF), (2) LOF or deleterious missense (LDM), and (3) coding, promotor, or enhancer variants.
Studies without sequencing data undertook single-variant analyses within each population group using their software of preference, and the same model described for TOPMed WGS analyses then provided summary statistics for central meta-analysis. Quality control of the study-specific single-variant GWAS summary statistics was undertaken using EasyQC31 using the following variant exclusion criteria: estimated MAC (MAC × imputation quality) <6, absolute effect size (β) >5, standard error >10, sample size <30, or imputation quality <0.30. Meta-analysis was completed using GWAMA32 with genomic control applied to each study but not to the meta-analysis results. Meta-analysis was completed (i) within each population group for studies with imputed data, (ii) within TOPMed WGS studies, and (iii) combined in a multipopulation mega-analysis that included results from (i) and (ii). Statistical significance was set at P < 5.0E-09.33
Conditional analysis, credible sets, and variance explained
For all genome-wide significant regions from the multipopulation mega-analysis, conditional analyses were undertaken using cojo-slct34 within GCTA35 (supplemental Figure 3). The linkage disequilibrium (LD) reference panel used all TOPMed WGS samples that contributed to this GWAS. The 95% credible sets were calculated for the same regions using the Wellcome Trust Case Control Consortium method.36 Percentage variance explained was estimated with summary-level data reported by GCTA35 for the 69 conditionally independent variants from the mega-analysis, using the approximation derived by Shim et al.37 See supplemental Methods for more details.
Functional annotation of fibrinogen-associated variants
Variant effect prediction
The Ensembl Variant Effect Predictor (VEP) (https://useast.ensembl.org/Tools/VEP)38 was used to determine nearest gene and top predicted consequence for each of the 69 conditionally independent variants and their LD proxies (as determined by r2 > 0.8 in TOP-LD European and/or African ancestry reference panels23,39). VEP was also used to annotate Combined Annotation Dependent Depletion (CADD) Phred40,41 and LoFtool42-predicted impact scores and additionally SIFT43 and PolyPhen44,45 scores for coding variants. InterPro was used to determine amino acid substitution.46
Overlap of fibrinogen signals and GWAS catalog associations
The same set of variants used for VEP annotation were queried, using rsID, against the National Human Genome Research Institute-European Bioinformatics Institute GWAS catalog v1.0.2, (downloaded 29 October 2022; https://www.ebi.ac.uk/gwas/downloads). Mapped traits corresponding to related quantitative measures were manually cataloged to broad categories (ie, liver enzymes, blood cell count and morphology traits, lipids, etc).
Colocalization
We performed colocalization to query gene-trait associations using fastENLOC47,48 with precomputed Genotype-Tissue Expression (GTEx) multiple-tissue expression quantitative trait loci (eQTL) annotations and fibrinogen GWAS posterior inclusion probabilities (PIPs) as inputs for each of the most relevant tissues: liver and whole blood. We considered a single-nucleotide polymorphism (SNP) level colocalization probability >0.1 or a regional colocalization probability (RCP) >0.5 as strong evidence of colocalization.
Transcription-wide association studies
A TWAS was performed using S-PrediXcan49 to identify associations between cis elements of gene expression and plasma levels of fibrinogen in mechanistically related tissues, namely, artery (aortic, coronary, or tibial), liver, and whole blood.50 Prebuilt prediction models, based on GTEx v8 multivariate adaptive shrinkage in R, were used to estimate variants’ weight on gene expression levels in chosen tissues.49-51 Because the reference models were created on the basis of the European-ancestry population, we limited the analysis to EUR-only GWAS results. S-PrediXcan results were evaluated using S-MultiXcan52 with significant TWAS signals determined using Bonferroni correction for the total number of genes across all models.
Fine mapping
TWAS fine mapping using FOCUS (Fine-mapping Of CaUsal gene Sets) was used to identify the likely causal gene under GWAS signals. FOCUS avoids false TWAS signals caused by coregulation and pleiotropic effects of variants at GWAS risk loci53 by modeling marginal TWAS z-scores of all genes in the same region considering variant LD correlations and tagged pleiotropic effects of variants on the trait. Given generated z-scores, the PIP for a gene to be causal is derived and then used to form a credible set of putative causal genes. GTEx v8 MASH-R models were used as the source of eQTL weights and the European-based PROCARDIS database as the reference for LD correlations. PIP ≥0.95 was used to identify a gene as putatively causal.
MVP PheWAS/Genetic risk score analysis
Genetic risk scores (GRSs) were derived using the independent variants identified by the GCTA analysis described above and as seen in supplemental Figure 3. Three scores were derived, (i) weighting by the variant β from the multipopulation mega-analysis, (ii) weighting by the variant β from the EUR-only meta-analysis, and (iii) weighting by the variant β from the AFR-only meta-analysis. GRSs were then standardized to standard deviation units. A PheWAS was performed for each of the 3 GRSs within EUR and AFR participants of the VA MVP,25 for International Classification of Diseases–based PheCodes54 with at least 500 cases. Logistic regression models were adjusted for age, sex, and the first 5 population-specific principal components, and statistical significance was determined on the basis of Bonferroni correction for the number of independent PheCodes (EUR PBonferroni = 5.18E-05; AFR PBonferroni = 7.25E-05) (see supplemental Data). Additional sensitivity analyses were completed by creating the GRS removing variants in (i) the FGG gene region or (ii) the fibrinogen gene cluster region.
Results
Baseline characteristics
WGS and fibrinogen measures were available for 32 572 individuals across 13 TOPMed studies. Imputed genetic data and fibrinogen measures were available for 131 340 individuals within 39 additional CHARGE or TOPMed studies. A total of 163 912 individuals were included in the multipopulation mega-analysis (Table 1). The mean fibrinogen value across studies was 3.32 g/L. Mean values were slightly higher in African-ancestry individuals compared with European (AFR = 3.59; EUR = 3.30), but the 95% confidence intervals overlapped in TOPMed participants (AFR = 1.67-5.56; EUR = 1.44-4.93). No difference in mean was observed between studies measuring functional fibrinogen using Clauss methods and those measuring total fibrinogen (Table 1). The heritability estimate from EUR TOPMed studies with WGS was 20%, which aligns with the lower end of estimates from family and twin studies.
Single-variant analysis and aggregate tests
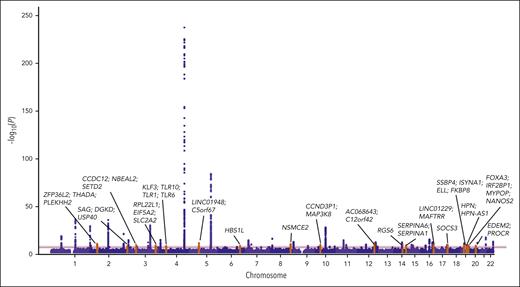
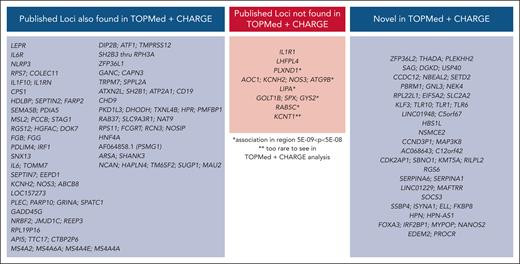
Multipopulation, single-variant mega-analysis identified 54 genetic loci associated with circulating fibrinogen (Figure 2; supplemental Table 1). Among these, 18 loci have not been identified in prior studies (Table 2; Figure 3). The TM6SF2 locus has been identified in a multiphenotype analysis leveraging fibrinogen and coronary artery disease variants,55 but not in a GWAS of fibrinogen alone. All 18 variants achieved nominal significance in a previously published analysis,18 and the direction of effect was consistent with our findings (Table 2). Associated regions contained an average of 21 SNPs in their 95% credible set (range, 1-93). Credible sets are summarized in supplemental Table 1, and full SNP lists are presented in supplemental Table 2. Approximate conditional analysis in GCTA revealed 6 loci harboring multiple conditionally distinct lead variants (Table 3; supplemental Table 3). At 4 loci with previously reported fibrinogen associations, FGG, PDLIM4, RPL22L1, and RAB37, we detected 7, 5, 3, and 2 distinct lead variants, respectively, exceeding the maximum from any prior report. supplemental Table 4 presents the LD between conditionally distinct lead variants and previously published top SNPs. Among newly associated loci, only SOCS3 and HPN contained multiple distinct lead variants.
Mega-analysis single-variant results. Orange peaks are novel associated regions and are labeled with the names of genes in the region. Red line indicates genome-wide significance (P < 5E-09), and blue line indicates suggestive association (P < 1E-07).
Mega-analysis single-variant results. Orange peaks are novel associated regions and are labeled with the names of genes in the region. Red line indicates genome-wide significance (P < 5E-09), and blue line indicates suggestive association (P < 1E-07).
Novel regions associated with fibrinogen in single-variant test
| SNP . | CHR . | POS (b38) . | EA . | NEA . | EAF . | β∗ . | SE . | P value . | Q P value . | No. (samples) . | Direction . | Gene region . | Top variant consequence . | de Vries et al 2015 . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β∗ . | P value . | R2 . | ||||||||||||||
| rs149290349 | 2 | 43 224 818 | A | G | 0.068 | 0.036 | 0.005 | 9.62E-12 | .102 | 163 060 | +−−+ | ZFP36L2; THADA; PLEKHH2 | Missense (ZFP36L2) | 0.008 | 9.70E-06 | 0.87 |
| rs60064064 | 2 | 233 460 824 | A | G | 0.089 | −0.028 | 0.005 | 3.90E-09 | .018 | 150 506 | −−+− | SAG; DGKD; USP40 | Intronic (DGKD) | −0.004 | 8.26E-03 | 0.90 |
| rs35053471 | 3 | 47 083 271 | A | T | 0.365 | 0.018 | 0.003 | 1.55E-10 | .873 | 163 912 | ++++ | CCDC12; NBEAL2; SETD2 | Intronic (SETD2) | 0.005 | 5.61E-07 | 0.86 |
| rs10936662 | 3 | 170 860 674 | A | T | 0.169 | 0.022 | 0.004 | 6.81E-10 | .408 | 163 912 | ++−+ | RPL22L1; EIF5A2; SLC2A2 | Intergenic | 0.005 | 2.18E-04 | 0.91 |
| rs4565087 | 4 | 38 761 482 | C | T | 0.369 | 0.017 | 0.003 | 1.97E-09 | .782 | 157 981 | ++−+ | KLF3; TLR10; TLR1; TLR6 | Intergenic | 0.005 | 2.75E-07 | 0.92 |
| rs157512 | 5 | 56 513 300 | C | T | 0.238 | 0.022 | 0.003 | 2.13E-12 | .140 | 163 912 | ++++ | LINC01948; C5orf67 | Intronic (C5orf67) | 0.004 | 4.66E-04 | 0.97 |
| rs9376090 | 6 | 135 090 090 | C | T | 0.235 | −0.019 | 0.003 | 1.63E-09 | .622 | 147 776 | −−−− | HBS1L | Intergenic | −0.005 | 4.97E-06 | 0.98 |
| rs6470350 | 8 | 125 315 303 | G | A | 0.511 | 0.019 | 0.003 | 1.91E-11 | .281 | 148 283 | ++−+ | NSMCE2 | Intronic | 0.004 | 6.15E-05 | 0.99 |
| rs1543725 | 10 | 30 409 944 | A | G | 0.408 | 0.018 | 0.003 | 1.35E-10 | .713 | 161 181 | ++++ | CCND3P1; MAP3K8 | Intergenic | 0.004 | 3.50E-05 | 0.94 |
| rs7298698 | 12 | 103 127 106 | C | T | 0.481 | 0.019 | 0.003 | 1.99E-12 | .463 | 161 688 | +−++ | AC068643; C12orf42 | Intronic (C12orf42) | 0.005 | 1.29E-07 | 0.99 |
| rs2239222 | 14 | 72 545 177 | G | A | 0.376 | −0.018 | 0.003 | 1.53E-10 | .002 | 147 776 | −−−− | RGS6 | Intronic (RGS6) | −0.005 | 4.79E-07 | 0.95 |
| rs17580 | 14 | 94 380 925 | A | T | 0.039 | 0.045 | 0.007 | 3.70E-10 | .291 | 163 770 | ++−+ | SERPINA6; SERPINA1 | Missense (SERPINA1) | 0.011 | 6.56E-07 | 0.70 |
| rs4334315 | 16 | 79 722 300 | T | A | 0.303 | 0.020 | 0.003 | 2.21E-11 | .723 | 163 912 | ++++ | LINC01229; MAFTRR | Intronic (LINC01229 and MAFTRR) | 0.005 | 2.84E-06 | 0.83 |
| rs11077357 | 17 | 78 347 520 | C | G | 0.767 | 0.023 | 0.004 | 1.35E-10 | .706 | 147 384 | ++−+ | SOCS3 | Intergenic | 0.006 | 7.41E-04 | 0.49 |
| rs7507218 | 19 | 18 525 110 | A | G | 0.220 | 0.020 | 0.003 | 1.04E-09 | .568 | 163 909 | ++++ | SSBP4; ISYNA1; ELL; FKBP8 | Intergenic | 0.004 | 6.68E-04 | 0.92 |
| rs1672981 | 19 | 35 056 147 | C | T | 0.916 | 0.034 | 0.005 | 4.07E-10 | .136 | 148 280 | +−++ | HPN; HPN-AS1 | Intronic (HPN) | 0.007 | 2.48E-04 | 0.95 |
| rs34934360 | 19 | 45 895 731 | G | A | 0.112 | −0.027 | 0.004 | 2.79E-10 | .996 | 150 507 | −−−− | FOXA3; IRF2BP1; MYPOP; NANOS2 | Intronic (MYPOP) | −0.007 | 4.37E-07 | 0.90 |
| rs1415771 | 20 | 35 146 690 | A | G | 0.471 | −0.017 | 0.003 | 1.20E-09 | .441 | 147 776 | −−−− | EDEM2; PROCR | Intronic (EDEM2) | −0.002 | 3.38E-02 | 1.00 |
| SNP . | CHR . | POS (b38) . | EA . | NEA . | EAF . | β∗ . | SE . | P value . | Q P value . | No. (samples) . | Direction . | Gene region . | Top variant consequence . | de Vries et al 2015 . | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β∗ . | P value . | R2 . | ||||||||||||||
| rs149290349 | 2 | 43 224 818 | A | G | 0.068 | 0.036 | 0.005 | 9.62E-12 | .102 | 163 060 | +−−+ | ZFP36L2; THADA; PLEKHH2 | Missense (ZFP36L2) | 0.008 | 9.70E-06 | 0.87 |
| rs60064064 | 2 | 233 460 824 | A | G | 0.089 | −0.028 | 0.005 | 3.90E-09 | .018 | 150 506 | −−+− | SAG; DGKD; USP40 | Intronic (DGKD) | −0.004 | 8.26E-03 | 0.90 |
| rs35053471 | 3 | 47 083 271 | A | T | 0.365 | 0.018 | 0.003 | 1.55E-10 | .873 | 163 912 | ++++ | CCDC12; NBEAL2; SETD2 | Intronic (SETD2) | 0.005 | 5.61E-07 | 0.86 |
| rs10936662 | 3 | 170 860 674 | A | T | 0.169 | 0.022 | 0.004 | 6.81E-10 | .408 | 163 912 | ++−+ | RPL22L1; EIF5A2; SLC2A2 | Intergenic | 0.005 | 2.18E-04 | 0.91 |
| rs4565087 | 4 | 38 761 482 | C | T | 0.369 | 0.017 | 0.003 | 1.97E-09 | .782 | 157 981 | ++−+ | KLF3; TLR10; TLR1; TLR6 | Intergenic | 0.005 | 2.75E-07 | 0.92 |
| rs157512 | 5 | 56 513 300 | C | T | 0.238 | 0.022 | 0.003 | 2.13E-12 | .140 | 163 912 | ++++ | LINC01948; C5orf67 | Intronic (C5orf67) | 0.004 | 4.66E-04 | 0.97 |
| rs9376090 | 6 | 135 090 090 | C | T | 0.235 | −0.019 | 0.003 | 1.63E-09 | .622 | 147 776 | −−−− | HBS1L | Intergenic | −0.005 | 4.97E-06 | 0.98 |
| rs6470350 | 8 | 125 315 303 | G | A | 0.511 | 0.019 | 0.003 | 1.91E-11 | .281 | 148 283 | ++−+ | NSMCE2 | Intronic | 0.004 | 6.15E-05 | 0.99 |
| rs1543725 | 10 | 30 409 944 | A | G | 0.408 | 0.018 | 0.003 | 1.35E-10 | .713 | 161 181 | ++++ | CCND3P1; MAP3K8 | Intergenic | 0.004 | 3.50E-05 | 0.94 |
| rs7298698 | 12 | 103 127 106 | C | T | 0.481 | 0.019 | 0.003 | 1.99E-12 | .463 | 161 688 | +−++ | AC068643; C12orf42 | Intronic (C12orf42) | 0.005 | 1.29E-07 | 0.99 |
| rs2239222 | 14 | 72 545 177 | G | A | 0.376 | −0.018 | 0.003 | 1.53E-10 | .002 | 147 776 | −−−− | RGS6 | Intronic (RGS6) | −0.005 | 4.79E-07 | 0.95 |
| rs17580 | 14 | 94 380 925 | A | T | 0.039 | 0.045 | 0.007 | 3.70E-10 | .291 | 163 770 | ++−+ | SERPINA6; SERPINA1 | Missense (SERPINA1) | 0.011 | 6.56E-07 | 0.70 |
| rs4334315 | 16 | 79 722 300 | T | A | 0.303 | 0.020 | 0.003 | 2.21E-11 | .723 | 163 912 | ++++ | LINC01229; MAFTRR | Intronic (LINC01229 and MAFTRR) | 0.005 | 2.84E-06 | 0.83 |
| rs11077357 | 17 | 78 347 520 | C | G | 0.767 | 0.023 | 0.004 | 1.35E-10 | .706 | 147 384 | ++−+ | SOCS3 | Intergenic | 0.006 | 7.41E-04 | 0.49 |
| rs7507218 | 19 | 18 525 110 | A | G | 0.220 | 0.020 | 0.003 | 1.04E-09 | .568 | 163 909 | ++++ | SSBP4; ISYNA1; ELL; FKBP8 | Intergenic | 0.004 | 6.68E-04 | 0.92 |
| rs1672981 | 19 | 35 056 147 | C | T | 0.916 | 0.034 | 0.005 | 4.07E-10 | .136 | 148 280 | +−++ | HPN; HPN-AS1 | Intronic (HPN) | 0.007 | 2.48E-04 | 0.95 |
| rs34934360 | 19 | 45 895 731 | G | A | 0.112 | −0.027 | 0.004 | 2.79E-10 | .996 | 150 507 | −−−− | FOXA3; IRF2BP1; MYPOP; NANOS2 | Intronic (MYPOP) | −0.007 | 4.37E-07 | 0.90 |
| rs1415771 | 20 | 35 146 690 | A | G | 0.471 | −0.017 | 0.003 | 1.20E-09 | .441 | 147 776 | −−−− | EDEM2; PROCR | Intronic (EDEM2) | −0.002 | 3.38E-02 | 1.00 |
CHR, chromosome; direction, effect direction from CHARGE EUR only, CHARGE AFR only, CHARGE HIS only, and TOPMed; EA, effect allele; EAF, effect allele frequency; NEA, noneffect allele; POS (b38), position in build 38; R2, imputation quality.
∗Absolute value of effect sizes (β) is not comparable between this study and de Vries et al due to differences in trait transformations before GWAS; deVries et al sample size = 120 246 (mean = 118 670, minimum = 102 767).
Comparison of mega-analysis single-variant results to previous publications.
Regions with multiple SNPs selected by GCTA (cojo-slct) using TOPMed multipopulation reference panel for LD
| Region . | Genes in region . | SNP . | CPTID . | refA . | freq_geno . | bJ . | bJ_se . | pJ . | LD_r . | SNP consequence . |
|---|---|---|---|---|---|---|---|---|---|---|
| 14 | RGS12; HGFAC; DOK7 | rs3748034 | chr4:3444364:G:T | T | 0.121 | 0.025 | 0.004 | 1.56E-10 | −0.086 | Missense (HGFAC) |
| rs16844401 | chr4:3447925:G:A | A | 0.059 | 0.045 | 0.005 | 4.01E-17 | 0.029 | Missense (HGFAC) | ||
| rs36205397 | chr4:3468877:A:G | G | 0.533 | 0.018 | 0.003 | 3.06E-10 | 0.000 | Intronic (DOK7) | ||
| 16 | FGB; FGG | rs2227401 | chr4:154565229:C:T | T | 0.176 | 0.093 | 0.003 | 2.14E-165 | −0.028 | Intronic (FGB) |
| rs6054 | chr4:154568456:C:T | T | 0.004 | −0.385 | 0.022 | 2.21E-67 | −0.011 | Missense (FGB) | ||
| rs2066874 | chr4:154608429:T:C | C | 0.008 | −0.294 | 0.030 | 4.02E-22 | −0.010 | Intronic (FGG) | ||
| rs148685782 | chr4:154611883:G:C | C | 0.003 | −0.805 | 0.024 | 2.88E-237 | −0.020 | Missense (FGG) | ||
| rs28577061 | chr4:154614588:T:C | C | 0.046 | −0.089 | 0.013 | 1.96E-12 | −0.030 | Intergenic | ||
| rs149748987 | chr4:154618106:T:C | C | 0.007 | −0.179 | 0.015 | 2.96E-32 | 0.090 | Intergenic | ||
| rs6536024 | chr4:154622217:T:C | C | 0.478 | −0.029 | 0.003 | 5.10E-25 | 0.000 | Intergenic | ||
| 18 | PDLIM4; SLC22A4; SLC22A5; IRF1; IL5; RAD50 | rs549687735 | chr5:132297674:G:A | A | 0.032 | 0.079 | 0.012 | 1.52E-10 | 0.157 | Intronic (SLC22A4) |
| rs145432904 | chr5:132454828:C:T | T | 0.030 | 0.082 | 0.007 | 3.99E-32 | −0.100 | Intronic (IRF1-AS1) | ||
| rs2706383 | chr5:132456710:G:A | A | 0.235 | −0.065 | 0.003 | 1.95E-80 | 0.000 | Intronic (IRF1-AS1) | ||
| rs2706339 | chr5:132464413:G:T | T | 0.236 | −0.065 | 0.003 | 9.23E-81 | −0.100 | Intronic (IRF1-AS1) | ||
| rs79480807 | chr5:132465298:G:A | A | 0.029 | 0.082 | 0.007 | 2.16E-32 | 0.000 | Intronic (IRF1-AS1) | ||
| 44 | RAB37; SLC9A3R1; NAT9 | rs4789661 | chr17:74429020:G:A | A | 0.635 | −0.030 | 0.004 | 3.49E-14 | 0.526 | Intergenic |
| rs1868057 | chr17:74720763:C:G | G | 0.628 | 0.039 | 0.004 | 7.41E-21 | 0.000 | Intronic (RAB37) | ||
| 45 | SOCS3 | rs11077357 | chr17:78347520:G:C | C | 0.761 | 0.023 | 0.004 | 9.10E-11 | −0.010 | Intergenic |
| rs938351 | chr17:78403323:C:T | T | 0.602 | 0.018 | 0.003 | 2.32E-10 | 0.000 | Intronic (PGS1) | ||
| 48 | HPN; HPN-AS1 | rs1672981 | chr19:35056147:T:C | C | 0.916 | 0.034 | 0.005 | 1.94E-10 | 0.021 | Intergenic (HPN) |
| rs2239943 | chr19:35539605:T:C | C | 0.597 | −0.017 | 0.003 | 1.76E-09 | 0.000 | Intergenic (GAPDHS) |
| Region . | Genes in region . | SNP . | CPTID . | refA . | freq_geno . | bJ . | bJ_se . | pJ . | LD_r . | SNP consequence . |
|---|---|---|---|---|---|---|---|---|---|---|
| 14 | RGS12; HGFAC; DOK7 | rs3748034 | chr4:3444364:G:T | T | 0.121 | 0.025 | 0.004 | 1.56E-10 | −0.086 | Missense (HGFAC) |
| rs16844401 | chr4:3447925:G:A | A | 0.059 | 0.045 | 0.005 | 4.01E-17 | 0.029 | Missense (HGFAC) | ||
| rs36205397 | chr4:3468877:A:G | G | 0.533 | 0.018 | 0.003 | 3.06E-10 | 0.000 | Intronic (DOK7) | ||
| 16 | FGB; FGG | rs2227401 | chr4:154565229:C:T | T | 0.176 | 0.093 | 0.003 | 2.14E-165 | −0.028 | Intronic (FGB) |
| rs6054 | chr4:154568456:C:T | T | 0.004 | −0.385 | 0.022 | 2.21E-67 | −0.011 | Missense (FGB) | ||
| rs2066874 | chr4:154608429:T:C | C | 0.008 | −0.294 | 0.030 | 4.02E-22 | −0.010 | Intronic (FGG) | ||
| rs148685782 | chr4:154611883:G:C | C | 0.003 | −0.805 | 0.024 | 2.88E-237 | −0.020 | Missense (FGG) | ||
| rs28577061 | chr4:154614588:T:C | C | 0.046 | −0.089 | 0.013 | 1.96E-12 | −0.030 | Intergenic | ||
| rs149748987 | chr4:154618106:T:C | C | 0.007 | −0.179 | 0.015 | 2.96E-32 | 0.090 | Intergenic | ||
| rs6536024 | chr4:154622217:T:C | C | 0.478 | −0.029 | 0.003 | 5.10E-25 | 0.000 | Intergenic | ||
| 18 | PDLIM4; SLC22A4; SLC22A5; IRF1; IL5; RAD50 | rs549687735 | chr5:132297674:G:A | A | 0.032 | 0.079 | 0.012 | 1.52E-10 | 0.157 | Intronic (SLC22A4) |
| rs145432904 | chr5:132454828:C:T | T | 0.030 | 0.082 | 0.007 | 3.99E-32 | −0.100 | Intronic (IRF1-AS1) | ||
| rs2706383 | chr5:132456710:G:A | A | 0.235 | −0.065 | 0.003 | 1.95E-80 | 0.000 | Intronic (IRF1-AS1) | ||
| rs2706339 | chr5:132464413:G:T | T | 0.236 | −0.065 | 0.003 | 9.23E-81 | −0.100 | Intronic (IRF1-AS1) | ||
| rs79480807 | chr5:132465298:G:A | A | 0.029 | 0.082 | 0.007 | 2.16E-32 | 0.000 | Intronic (IRF1-AS1) | ||
| 44 | RAB37; SLC9A3R1; NAT9 | rs4789661 | chr17:74429020:G:A | A | 0.635 | −0.030 | 0.004 | 3.49E-14 | 0.526 | Intergenic |
| rs1868057 | chr17:74720763:C:G | G | 0.628 | 0.039 | 0.004 | 7.41E-21 | 0.000 | Intronic (RAB37) | ||
| 45 | SOCS3 | rs11077357 | chr17:78347520:G:C | C | 0.761 | 0.023 | 0.004 | 9.10E-11 | −0.010 | Intergenic |
| rs938351 | chr17:78403323:C:T | T | 0.602 | 0.018 | 0.003 | 2.32E-10 | 0.000 | Intronic (PGS1) | ||
| 48 | HPN; HPN-AS1 | rs1672981 | chr19:35056147:T:C | C | 0.916 | 0.034 | 0.005 | 1.94E-10 | 0.021 | Intergenic (HPN) |
| rs2239943 | chr19:35539605:T:C | C | 0.597 | −0.017 | 0.003 | 1.76E-09 | 0.000 | Intergenic (GAPDHS) |
bJ, joint β; bJ_se, standard error of joint β; CPTID, chromosome:position (b38):allele1: allele 2; freq_geno, frequency of effect allele in reference sample; LD_r, linkage disequilibrium; pJ, joint P value; refA, reference/effect allele.
Among the 69 conditionally distinct lead variants associated with fibrinogen, 5 variants (rs10936662, rs28577061, rs11077357, rs7507218, and rs1672981) have a minor allele at least 10 percentage points more frequent in African ancestral populations compared with European (supplemental Table 3). At the FGG locus, GCTA identified a conditionally distinct lead variant (rs28577061), which has not been associated with fibrinogen in prior studies and is common in AFR but extremely rare or absent in other TOPMed populations (MAFAFR = 0.1801; MAFEUR = 0.008). At 4 loci, RPL22L1, HPN, SOCS3, and SSPB4, the primary lead variants are common in all assessed populations, but at increased frequency in African ancestry populations. Together, the 69 independent variants discovered increased the phenotypic variance explained for circulating fibrinogen across populations from 3.7%18 to 4.8%, with the fibrinogen gene cluster alone explaining 2.2%. Effect sizes were highly correlated between studies that measured functional fibrinogen and those that measured total fibrinogen (supplemental Figure 4; supplemental Table 5).
Aggregate tests using low-frequency and rare TOPMed WGS variants yielded associations in the fibrinogen gene cluster region: FGG was significant when aggregating LOF and LDM variants, whereas FGA was only significant when aggregating LOF variants and FGB with LDM variants. No genes were significant when aggregating all low-frequency and rare variants in the coding, promotor, or enhancer regions (supplemental Table 6). A list of variants in this region included in the aggregation tests is presented in supplemental Table 7.
Variant annotation
To characterize each genetic signal associated with fibrinogen, we queried all conditionally distinct variants and their LD proxies (r2 > 0.8 in TOPMed-based EUR and/or AFR ancestry reference panels),23,39 in multiple publicly available data sets. Most signals we identified contained previously reported lead variants for liver-enzyme measures, lipid measures, and/or blood-cell traits. Notably, 23 signals contained a previously reported lead variant for C-reactive protein (CRP).
VEP38 annotation shows that 18 signals contained at least 1 missense variant. Additionally, 13 signals contained variants with CADD PHRED scores exceeding 20, indicating the variant is in the top 1% of predicted deleterious mutations. SERPINA1, ZFP36L2, and TLR10 stand out as newly associated loci in this category. We also observed that several signals overlap potential regulatory regions in liver. A total of 52 signals (45 loci) contain at least 1 variant mapping to a “consensus” region of open chromatin in liver tissue samples, 42 signals overlap a GeneHancer regulatory element reported active in HepG2 hepatocytes, 5 signals harbor previously reported liver chromatin accessibility quantitative trait loci (QTL) variants, 6 signals hold GTEx liver expression QTL, and the primary signal in HPN contains a GTEx liver splice QTL. A summary of these annotations for each signal can be found in supplemental Table 8 and full results in supplemental Table 9.
Colocalization
Variant-level colocalization analysis in fastENLOC identified 153 variant-tissue pairs with evidence (SNP-level colocalization probability >0.1) for a shared genetic basis with GTEx eQTLs for 93 distinct genes in aortic artery, tibial artery, coronary artery, liver, and/or whole blood (supplemental Table 10). Regional analysis in the same set of tissues found 46 region-tissue pairs, implicated in regulation of 41 distinct genes, with evidence for statistical colocalization with fibrinogen regions (RCP > 0.5) (supplemental Table 11). We note that 5 of the 6 fibrinogen signals harboring sentinel GTEx liver eQTL variants statistically colocalize with the eQTL signals in fastENLOC, whereas the PLEC eQTL signal was at the significance threshold (RCP = 0.5).
Transcriptome-wide association study
Gene-tissue pairs across 5 tissues (aortic, coronary, or tibial artery, whole blood, and liver) were included in MetaXcan TWAS analyses for fibrinogen (supplemental Figure 5). Genetically determined expressions of 64 gene-tissue pairs were significantly associated with fibrinogen levels after Bonferroni correction and filtering for models with at least 2 contributing SNPs (supplemental Table 12). A total of 15 gene-tissue pairs identified by TWAS were further prioritized via FOCUS fine-mapping (supplemental Table 12). TNKS was the only gene prioritized by both TWAS and fine-mapping in liver analyses. Notably, this gene was further implicated in fibrinogen regulation through variant and region-based liver expression colocalization analysis (supplemental Table 11). In whole blood analyses, TWAS and fine-mapping prioritized AFT1, C5orf56, FGB, MS4A4E, and SLC22A5. Of these, AFT1 was further supported by variant-level colocalization analysis (supplemental Table 10).
Phenome-wide association study
Three fibrinogen GRSs were tested for association with 1426 PheCodes in EUR and 1061 PheCodes in AFR participants of the VA MVP (supplemental Table 13). Each score reached Bonferroni significance with multiple thrombotic and inflammatory disease phenotypes (nEUR = 13 PheCodes, nAFR = 1 PheCode; Table 4; supplemental Table 14). Top associations included venous thromboembolism and gout.
Association of TOPMed/CHARGE fibrinogen PRS with PheCodes in MVP∗
| Trait information . | MVP EUR . | MVP AFR . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PheCode . | Disease category . | Description . | No. of cases/controls . | PRS (MEGA) . | PRS (EUR) . | No. of cases/controls . | PRS (MEGA) . | PRS (AFR) . | ||||
| OR . | P value . | OR . | P value . | OR . | P value . | OR . | P value . | |||||
| phe_452 | Circulatory system | Other venous embolism and thrombosis | 26 792/421 526 | 0.952 | 2.26E-15† | 0.952 | 1.35E-15† | 7585/111 531 | 0.977 | 5.25E-02 | 0.972 | 1.76E-02‡ |
| phe_452_2 | Circulatory system | Deep vein thrombosis | 14 944/438 829 | 0.941 | 9.78E-14† | 0.941 | 6.88E-14† | 4498/116 209 | 0.964 | 1.60E-02‡ | 0.962 | 1.00E-02§ |
| phe_274_1 | Endocrine or metabolic | Gout | 42 682/406 941 | 1.031 | 2.96E-09† | 1.032 | 1.82E-09† | 14 511/104 430 | 1.022 | 1.51E-02‡ | 1.012 | 1.98E-01 |
| phe_274 | Endocrine or metabolic | Gout and other crystal arthrop | 44 888/402 674 | 1.030 | 4.02E-09† | 1.031 | 2.57E-09† | 14 941/103 648 | 1.022 | 1.43E-02‡ | 1.012 | 1.92E-01 |
| phe_681 | Dermatologic | Superficial cellulitis and abscess | 74 338/335 942 | 0.979 | 1.38E-07† | 0.979 | 1.80E-07† | 19 580/88 222 | 0.989 | 1.51E-01 | 0.988 | 1.38E-01 |
| phe_1089 | Other | Acquired absence of limb | 5 466/454 009 | 0.940 | 3.54E-06† | 0.940 | 3.83E-06† | 1675/120 554 | 0.998 | 9.50E-01 | 1.010 | 6.93E-01 |
| phe_681_5 | Dermatologic | Cellulitis and abscess of leg, except foot | 20 789/421 257 | 0.970 | 1.37E-05† | 0.970 | 1.35E-05† | 3302/115 194 | 1.000 | 9.85E-01 | 1.004 | 8.38E-01 |
| phe_280 | Hematopoietic | Iron-deficiency anemias | 35 686/404 586 | 1.024 | 1.94E-05† | 1.024 | 3.20E-05† | 14 819/100 241 | 1.038 | 3.97E-05† | 1.034 | 1.71E-04§ |
| phe_707_2 | Dermatologic | Chronic ulcer of leg or foot | 18 477/432 947 | 0.969 | 2.00E-05† | 0.969 | 2.37E-05† | 4470/115 706 | 0.994 | 6.73E-01 | 0.993 | 6.54E-01 |
| phe_286_81 | Hematopoietic | Primary hypercoagulable state | 2 498/457 232 | 0.921 | 2.51E-05† | 0.920 | 2.03E-05† | 529/121 823 | 0.957 | 3.11E-01 | 0.947 | 2.07E-01 |
| phe_286_2 | Hematopoietic | Encounter for long-term (current) use of anticoagulants | 42 101/409 581 | 0.979 | 3.04E-05† | 0.978 | 2.00E-05† | 7617/112 768 | 0.963 | 1.49E-03§ | 0.960 | 5.44E-04§ |
| phe_286 | Hematopoietic | Coagulation defects | 12 517/436 641 | 0.963 | 3.30E-05† | 0.963 | 3.18E-05† | 2804/116 917 | 0.936 | 4.47E-04§ | 0.934 | 3.01E-04§ |
| phe_286_8 | Hematopoietic | Hypercoagulable state | 2 691/456 755 | 0.925 | 3.52E-05† | 0.925 | 3.18E-05† | 586/121 682 | 0.964 | 3.70E-01 | 0.952 | 2.35E-01 |
| Trait information . | MVP EUR . | MVP AFR . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PheCode . | Disease category . | Description . | No. of cases/controls . | PRS (MEGA) . | PRS (EUR) . | No. of cases/controls . | PRS (MEGA) . | PRS (AFR) . | ||||
| OR . | P value . | OR . | P value . | OR . | P value . | OR . | P value . | |||||
| phe_452 | Circulatory system | Other venous embolism and thrombosis | 26 792/421 526 | 0.952 | 2.26E-15† | 0.952 | 1.35E-15† | 7585/111 531 | 0.977 | 5.25E-02 | 0.972 | 1.76E-02‡ |
| phe_452_2 | Circulatory system | Deep vein thrombosis | 14 944/438 829 | 0.941 | 9.78E-14† | 0.941 | 6.88E-14† | 4498/116 209 | 0.964 | 1.60E-02‡ | 0.962 | 1.00E-02§ |
| phe_274_1 | Endocrine or metabolic | Gout | 42 682/406 941 | 1.031 | 2.96E-09† | 1.032 | 1.82E-09† | 14 511/104 430 | 1.022 | 1.51E-02‡ | 1.012 | 1.98E-01 |
| phe_274 | Endocrine or metabolic | Gout and other crystal arthrop | 44 888/402 674 | 1.030 | 4.02E-09† | 1.031 | 2.57E-09† | 14 941/103 648 | 1.022 | 1.43E-02‡ | 1.012 | 1.92E-01 |
| phe_681 | Dermatologic | Superficial cellulitis and abscess | 74 338/335 942 | 0.979 | 1.38E-07† | 0.979 | 1.80E-07† | 19 580/88 222 | 0.989 | 1.51E-01 | 0.988 | 1.38E-01 |
| phe_1089 | Other | Acquired absence of limb | 5 466/454 009 | 0.940 | 3.54E-06† | 0.940 | 3.83E-06† | 1675/120 554 | 0.998 | 9.50E-01 | 1.010 | 6.93E-01 |
| phe_681_5 | Dermatologic | Cellulitis and abscess of leg, except foot | 20 789/421 257 | 0.970 | 1.37E-05† | 0.970 | 1.35E-05† | 3302/115 194 | 1.000 | 9.85E-01 | 1.004 | 8.38E-01 |
| phe_280 | Hematopoietic | Iron-deficiency anemias | 35 686/404 586 | 1.024 | 1.94E-05† | 1.024 | 3.20E-05† | 14 819/100 241 | 1.038 | 3.97E-05† | 1.034 | 1.71E-04§ |
| phe_707_2 | Dermatologic | Chronic ulcer of leg or foot | 18 477/432 947 | 0.969 | 2.00E-05† | 0.969 | 2.37E-05† | 4470/115 706 | 0.994 | 6.73E-01 | 0.993 | 6.54E-01 |
| phe_286_81 | Hematopoietic | Primary hypercoagulable state | 2 498/457 232 | 0.921 | 2.51E-05† | 0.920 | 2.03E-05† | 529/121 823 | 0.957 | 3.11E-01 | 0.947 | 2.07E-01 |
| phe_286_2 | Hematopoietic | Encounter for long-term (current) use of anticoagulants | 42 101/409 581 | 0.979 | 3.04E-05† | 0.978 | 2.00E-05† | 7617/112 768 | 0.963 | 1.49E-03§ | 0.960 | 5.44E-04§ |
| phe_286 | Hematopoietic | Coagulation defects | 12 517/436 641 | 0.963 | 3.30E-05† | 0.963 | 3.18E-05† | 2804/116 917 | 0.936 | 4.47E-04§ | 0.934 | 3.01E-04§ |
| phe_286_8 | Hematopoietic | Hypercoagulable state | 2 691/456 755 | 0.925 | 3.52E-05† | 0.925 | 3.18E-05† | 586/121 682 | 0.964 | 3.70E-01 | 0.952 | 2.35E-01 |
OR, odds ratio per SD increase in the PRS.
∗Analyses were only run if there were >500 cases or controls for a specific PheCode. Bonferroni threshold was determine separately in each ancestry group. EUR had 1426 PheCodes analyzed with an estimated 965 independent traits; therefore, Bonferroni = 0.05/965 = 5.18E-05. AFR had 1061 PheCodes analyzed with an estimated 690 independent traits; therefore, Bonferroni = 0.05/690 = 7.25E-05.
P ≤ Bonferroni threshold.
01 ≤ P < Bonferroni.
05 ≤ P < .01.
To further assess which variants within the GRS may drive individual associations, we queried each variant against the significant PheCode (supplemental Table 15). As anticipated, for “Circulatory System” and “Hematopoietic” PheCodes, most of the signal came from variants in the fibrinogen gene cluster, whereas significant variants across the GRS were associated with gout and dermatologic traits. Sensitivity analyses with PRSs removing variants (i) in the FGG gene region (associated with γ prime fibrinogen) or (ii) in the full fibrinogen gene cluster did not substantially change the odds ratio, but did impact the significance (supplemental Table 16). PheCodes relating to gout (phe_274, phe_274_1) and superficial cellulitis and abscess (phe_1089) became more significant, whereas PheCodes relating to coagulation defects and hypercoagulable state (phe_286, phe_286_8, and phe_286_81) became less significant.
Magnetic resonance (MR) analysis of fibrinogen and gout failed to provide evidence for a causal effect of fibrinogen levels on gout, and multivariable MR analysis incorporating CRP levels to account for possible confounding was inconclusive (supplemental Table 17).
Discussion
Herein, we used WGS and genotype data from diverse participants to identify 69 conditionally distinct genetic variants across 54 loci associated with circulating fibrinogen. Our results corroborate previous reports that fibrinogen is highly polygenic and advance the field by identifying new variant associations, including rare variants and variants most prevalent in underrepresented populations. On the basis of results of in silico characterization, we suggest some of the new genetic regulators we identified may act directly to regulate coagulation factors (such as factor VII and factor XI), whereas others may impact fibrinogen broadly through pathways altering liver metabolism, inflammation, and immune function, as reflected in the broad overlap between fibrinogen-associated signals and prior analyses for liver-related traits and blood and immune-cell counts.
Population-differentiated variants among new associations
We identified several novel genetic associations driven by variants with higher allele frequencies in non-European populations, validating that our approach improved detection of putative genetic regulators most common in underrepresented populations. To our knowledge, we identified the first common variant-fibrinogen association driven almost entirely by African ancestry participants at an intergenic region near FGG. Previous African ancestry driven variants were rare in African and not present in European populations.19 Additionally, expanded representation of both African and European populations allowed us to detect 4 new signals of small effect, led by common variants substantially more frequent in those with African ancestry. Detection of population-differentiated associations indicates that for variants with higher predicted effects, such as those within the fibrinogen gene cluster, expanded sample sizes are now reaching power to detect variant associations driven by underrepresented populations. We further identified 4 common European-driven signals that are rare or uncommon in African ancestry populations, suggesting that genomic signals for circulating fibrinogen are not saturated in any ancestral population.56,57 This is also reflected by the relatively modest variance explained (4.8%) vs estimates of total fibrinogen heritability (30%-50%).12,13,17 Increased diversity and larger sample sizes are still needed to help close this gap.
Newly identified potential coagulation pathway interactors
Three novel regions (SERPINA1, ZFP36L2, and TLR10) harbor deleterious missense variants in genes with plausible connections to coagulation and/or inflammation. In the SERPINA1 gene, we identified a rare missense variant (rs17580) predicted to be highly deleterious (CADD Phred score = 32). SERPINA1 encodes α-1-antitrypsin, which protects other proteins and tissues from serine protease degradation via direct binding inhibition.58 Like fibrinogen, α-1-antitrypsin is synthesized in hepatocytes in response to IL-6 and IL-1 inflammatory pathways.59 Severe α-1-antitrypsin deficiency is associated with liver dysfunction and emphysema related to a lack of inhibition of neutrophil elastase.60 The association of SERPINA1 with fibrinogen levels may reflect α-1-antitrypsin involvement in inflammatory pathways, or a more direct relationship between these proteins. Although in vitro assays have suggested α-1-antitrypsin can bind to plasma fibrinogen and be incorporated into fibrin networks,58 little is known about the functional implications of these interactions and whether it also influences fibrin(ogen) degradation.
Zinc Finger Protein (ZFP)36L2 and toll-like receptor 10 (TLR10) are broadly linked to inflammatory pathways. Although ZFP36L2 is largely uncharacterized, the ZFP36L2 missense variant we observed has previously been associated with blood cell count and morphology traits,61-64 and studies in mice show ZFP36L2 regulates blood cell development and adipogenesis.65 Proteins of the ZFP36 family regulate transcript abundance through binding AU-rich elements to target transcripts for decay.66 Notably, ZFP36 (tristetraprolin) family proteins are known to modulate transcript abundance of the inflammatory cytokine tumor necrosis factor-α,67 which induces both the coagulation cascade and complement system.68,69TLR10 encodes a TLR protein. Although TLR activation generally contributes to inflammation, immune responses, and activation of coagulation cascades, little is known about TLR10 specifically.70 Although TLR10 may exert a protective effect against inflammation,71,72 more work is needed to elucidate mechanistic links between this protein and coagulation.
Putative fibrinogen regulatory signals
Although the missense variants highlight intriguing new gene targets for future functional studies, most signals we identified are driven by common noncoding variants with small effect sizes. Many of these noncoding signals overlap HepG2-active regulatory regions, and a subset of signals overlap liver chromatin accessibility, expression, and/or splice QTL signals, suggesting these variants may exert subtle regulatory effects on circulating fibrinogen levels. Notably, at 2 newly associated loci, HPN and SOCS3, we identified 2 conditionally distinct variants, each with distinct functional annotations.
At the HPN locus, the primary signal, which maps to an intronic region of HPN, contains a liver splice QTL for HPN. HPN encodes hepsin, a type II transmembrane serine protease with “enhanced” expression in the liver (according to the Human Protein Atlas), believed to function in macromolecular metabolism.73 Interestingly, studies in zebrafish and mammalian cell lines suggest hepsin activates coagulation factor VII.74,75 Although mouse studies have not conclusively demonstrated hepsin’s involvement in coagulation, these studies have established hepsin’s involvement in regulating liver metabolism via hepatocyte growth factor and Met signaling pathways.73,76-78 The secondary signal at this locus maps to an intergenic region and contains a liver eQTL for a nearby gene, TMEM147. TMEM147, which was also prioritized in liver, whole blood, and aortic artery TWAS analyses, encodes a widely expressed endoplasmic reticulum transmembrane protein implicated in various metabolic processes, including calcium transport.79 Although it is unclear whether the 2 signals in this region influence fibrinogen through altered splicing and expression patterns of HPN and TMEM147 in the liver, these signals provide a compelling starting point for future functional studies.
Similarly, among the 2 independent, common-variant signals in the SOCS3, PGS1 locus on chromosome 17, the secondary signal houses a previously identified sentinel variant for altered chromatin accessibility in liver tissue. This variant is of particular interest given its proximity to SOCS3, a “suppressor of cytokine signaling” known to act upstream of IL-6 in the acute-phase response pathway that induces fibrinogen.2 It is possible that the secondary signal at this locus is capturing genetic variation that modulates the accessibility of the SOCS3 gene, and potentially other nearby genes, in liver or other tissues, leading to downstream impacts on fibrinogen levels. SOCS3 methylation was recently associated with circulating fibrinogen levels in an epigenome-wide association study in the CHARGE consortium80 with the associated probe within 11 kb of our best SOCS3 GWAS SNP, reinforcing the idea that chromatin accessibility may be at play.
PheWAS
Results of the PheWAS in MVP yielded expected associations with venous thromboembolism and several hematopoietic traits. However, the direction of the association within bleeding and thrombosis phenotypes was often opposite to the direction we expected given total fibrinogen’s procoagulant function (ie, typically increased genetically predicted fibrinogen is associated with decreased risk of bleeding and increased risk of thrombosis). Although seemingly paradoxical, this finding is in line with what has been observed in patients with congenital fibrinogen deficiency. This rare, life-threatening bleeding disorder results from a complete or partial loss of fibrinogen, and/or the presence of dysfunctional fibrinogen.2,81 In patients with complete loss of fibrinogen, not only has increased bleeding (from loss of fibrin formation) been reported, but also increased risk of thrombosis (hypothesized to result from loss of the anticoagulant properties of the γ' isoform), demonstrating complex and sometimes competing effects of fibrinogen isoforms on coagulation biology.82 A previous MR study observed an inverse association between total fibrinogen levels with venous thromboembolism risk.9 We tested the hypothesis that this inverse association in our PheWAS was driven by the alternatively spliced γ prime fibrinogen isoform, which has established anticoagulant properties,83 but removing the FGG locus SNPs from the tested PRS did not alter the direction of effect we observed. Our hypothesis is thus that genetically driven increases in total fibrinogen are accompanied by increases in γ prime fibrinogen, and that the antithrombotic effect of γ prime fibrinogen compensates for the prothrombotic effect of other isoforms or of total fibrinogen.
Another unexpected finding from the PheWAS was a positive association between genetically predicted total fibrinogen and gout. Interestingly, this finding is in line with emerging literature, which has shown that patients with active gout have increased thrombin generation markers.84,85 Our MR analysis did not support a causal association of fibrinogen on gout, and multivariable Mendelian randomization analysis to investigate if inflammation plays a profound role in the association between fibrinogen and gout (using CRP as a proxy) was inconclusive. Given the extensive overlap between our signals and previously reported GWAS signals for phenotypes reflecting liver health and inflammation, such as liver enzyme and lipid measures, we suggest that this association may be capturing variant impacts on broader liver metabolic pathways.
Conclusion
In conclusion, we identified 18 novel loci, collectively harboring 20 distinct variants, associated with fibrinogen measure. We report the first common African-driven fibrinogen association, and several additional associated variants with population-differentiated genetic architecture. Furthermore, we demonstrate overlap between these signals and liver regulatory elements, as well as GWAS phenotypes reflecting altered liver metabolism and inflammation. Future studies investigating coregulation and epistatic effects will likely provide new insight on the shared genetic architecture, and biological interplay, of hemostasis and inflammation.
Acknowledgments
The authors gratefully acknowledge the studies and participants who provided biological samples and data for TOPMed.
Molecular data for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Institutes of Health (NIH), National Heart, Lung, and Blood Institute (NHLBI). See the TOPMed Omics Support Table in the supplemental Data for study-specific omics support information. Core support, including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering, was provided by the TOPMed Informatics Research Center (NIH, NHLBI grant 3R01HL-117626-02S1; contract HHSN268201800002I). Core support, including phenotype harmonization, data management, sample-identity quality control, and general program coordination, was provided by the TOPMed Data Coordinating Center (NIH, NHLBI grants R01HL-120393; U01HL-120393; contract HHSN268201800001I). CHARGE is supported by NIH, NHLBI grant R01HL-105756, and the CHARGE and TOPMed Hemostasis Working Groups by NIH, NHLBI grants R01HL-134894, R01HL-139553, and R01HL-141291. This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by award I01-BX004821. The study was supported by the Novo Nordisk Foundation (grant NNF18CC0034900). D.-A.T. was supported by the EPIDEMIOM-VT Senior Chair of Excellence from the University of Bordeaux initiative of Excellence. M.S.-L. is supported by a Miguel Servet contract from the ISCIII Spanish Health Institute (CPII22/00007) and cofinanced by the European Social Fund. Additional study-specific funding and acknowledgments can be found in the supplemental Data. The Genotype-Tissue Expression Project was supported by the Common Fund of the NIH Office of the Director, and by the NIH, National Cancer Institute, NIH, National Human Genome Research Institute, NIH, NHLBI, NIH, National Institute on Drug Abuse, NIH, National Institute of Mental Health, and NIH, National Institute of Neurological Disorders and Stroke.
This publication does not represent the views of the Department of Veteran Affairs, the US National Institutes of Health, the National Heart, Lung, and Blood Institute, the US Department of Health and Human Services, or the US government.
Authorship
Contribution: L.M.R., J.A.B., L.A., D.I.C., M.-H.C., J.P.L., O. Polasek, J.I.R., E.d.G., J.-F.D., L.E., N.F., M.G., A.L., S.S.R., F.R.R., I.R., J.C.S., L.C.B., A.B., H.C., K.C., J.E.C., M.D., P.E., M.N., V.O., B.M.P., J.M.S., D.J.S., A.v.H., H.W., W.F.W., Y.B.-S., J.B., D.B., S.R.C., J.G.E., E.F., C.H., M.A.I., J.W.J., S.L.R.K., L.A.L., R.A.M., B.D.M., D.O.M.-K., P.-E.M., P.P.P., A.R., P.M.R., T.D.S., N.J.W., J.F.W., D.-A.T., A.D.J., P.S.d.V., A.C.M., and N.L.S. participated in the design and/or funding of the contributing studies; J.E.H., L.M.R., L.R.Y., L.A., L.F.B., R.P.B., G.D.C., J.L., J.P.L., R.L.-G., M.M., O. Polasek, M.S., P.S., M.H.C., P.C., E.d.G., N.F., M.G., A.G., H.G., T.H., T.K., J.L., A.L., S.N., F.R.R., I.R., K.A.R., J.C.S., F.J.A.v.R., H.W., W.Z.A.B., B.E.C., K.C., J.D.C., M.P.M.d.M., M.D., P.E., C.F., N.G., S.H.E., I.K., M.N., J.R.O., V.O., B.M.P., K.R., J.A.S., D.J.S., A.v.H., G.W., Y.B.-S., J.B., D.B., S.R.C., J.G.E., E.F., T.H., S.L.R.K., L.A.L., B.D.M., P.-E.M., O. Pedersen, S.R., P.M.R., E.K.S., N.J.W., J.F.W., P.S.d.V., M.S.-L., A.C.M., and N.L.S. participated in phenotype data acquisition and/or quality control; L.R.Y., L.A., G.D.C., D.I.C., M.-H.C., D.B.E., J.H., J.-J.H., M.E.K., N.-Q.L., J.L., R.L.-G., M.M., A.M.-P., A.R., B.A.T.R., M.S., P.S., S.T., S.W., M.Z., P.A., M.H.C., G.D., E.d.G., J.-F.D., H.G., T.O.K., J.L., A.L., S.N., F.P.-V., F.R.R., K.A.R., F.J.A.v.R., H.W., W.Z., B.E.C., J.E.C., C.F., N.G., S.H.E., C.M., J.R.O., V.O., B.M.P., K.R., J.A.S., G.W., J.B., D.B., A.D., J.G.E., E.F., M.F., T.H., C.H., J.W.J., S.L.R.K., R.A.M., B.D.M., D.O.M.-K., P.-E.M., O. Pedersen, S.R., P.M.R., E.K.S., U.V., J.F.W., D.-A.T., A.D.J., M.S.-L., and A.C.M. participated in genotype data acquisition and/or quality control; J.E.H., J.N., J.H., A.S.H., L.R.Y., J.A.B., F.T., L.F.B., G.D.C., M.-H.C., D.B.E., M.G., J.-J.H., M.E.K., N.-Q.L., J.P.L., R.L.-G., J.L., A.M., M.M., R.E.M., A.M.-P., A.R., B.A.T.R., M.S., P.S., S.T., S.W., M.Z., P.A., G.D., G.E.D., M.G., P.K.J., S.N., R.N., F.P.-V., K.A.R., M.R.B., J.S.F., C.F., X.G., C.K., J.R.O., D.B., A.D., C.H., R.A.M., B.D.M., A.R., U.V., J.Y., D.-A.T., A.D.J., P.S.d.V., M.S.-L., A.C.M., and N.L.S. participated in data analysis and/or interpretation; and J.E.H., J.N., J.H., P.S.d.V., M.S.-L., A.C.M., and N.L.S. participated in drafting the initial manuscript; and all authors participated in critical review of the manuscript.
Conflict-of-interest disclosure: L.R. is a consultant for the TOPMed Administrative Coordinating Center (through WeStat). R.L.-G. is a part-time consultant for Metabolon, Inc. M.C. has received grant support from Bayer. B.P. serves on the Steering Committee of the Yale Open Data Access Project, funded by Johnson & Johnson. In the past 3 years, E.K.S. has received grant support from Bayer. The remaining authors declare no competing financial interests.
A complete list of the members of the VA Million Veteran Program and the National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) Consortium appears in the supplemental Data.
Correspondence: Jennifer E. Huffman, 2 Ave de Lafayette, ATTN: VA MAVERIC, Boston, MA 02110; email: jennifer.huffman2@va.gov.
References
Author notes
J.E.H., J.N., and J.H. are joint first authors.
P.S.d.V., M.S.-L., A.C.M. and N.L.S. are joint last authors.
Whole-genome sequencing and selected phenotype data for TOPMed studies are available through dbGaP and the National Heart, Lung, and Blood Institute BioData Catalyst cloud platform (see https://topmed.nhlbi.nih.gov/topmed-data-access-scientific-community for detailed instructions). dbGaP accession numbers for studies specifically contributing to this study can also be found in the supplemental Data (“TOPMed Omics Support Table”). Full genome-wide association study summary statistics for this analysis are available through the “CHARGE Consortium Summary Results from Genomics Studies” dbGaP page (accession phs000930) or by contacting the corresponding author, Jennifer E. Huffman (jennifer.huffman2@va.gov).
The data used for the analyses described in this article were obtained from the Gentotype-Tissue Expression Portal on 29 October 2022.
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal