In this issue of Blood, Millman et al1 identify the α-ketoglutarate dehydrogenase (α-KGDH) complex in mitochondrial metabolism as a critical metabolic node and potential therapeutic vulnerability in acute myeloid leukemia (AML).
AML is a heterogenous group of hematologic cancers characterized by distinct metabolic requirements to sustain proliferation, survival, and redox homeostasis.2 AML cells reprogram their metabolism, relying on pathways such as glycolysis, oxidative phosphorylation (OXPHOS), amino acid metabolism, and lipid metabolism to generate biomolecules and meet high bioenergetic demands. These metabolic dependencies have been exploited as therapeutic targets for the development of metabolism-focused strategies,3 including inhibitors of oncogenic isocitrate dehydrogenase mutations and antagonists targeting OXPHOS, glutaminase, dihydroorotate dehydrogenase, and the anti-apoptotic B-cell leukemia/lymphoma 2 protein (eg, venetoclax). While single-agent or combinatorial therapies targeting metabolism show promise, challenges such as disease heterogeneity, relapse, and resistance highlight the need for personalized treatment approaches.
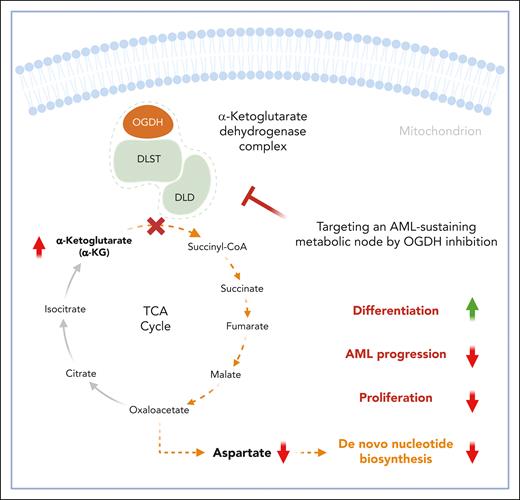
In this study, Millman et al describe α-KGDH as a critical metabolic node driving AML progression. By analyzing genome-wide CRISPR-Cas9 screening data involving over 1000 cancer cell lines, including 24 AML lines,4 they identified the genes encoding components of the α-KGDH complex—2-oxoglutarate dehydrogenase (OGDH), dihydrolipoyl succinyltransferase (DLST), and dihydrolipoyl dehydrogenase (DLD)—as specific, genotype-independent dependencies in AML (see figure). Given the role of α-KGDH in α-ketoglutarate (α-KG) production, cell fate regulation, and tumor suppression,5,6 these findings uncover a previously unrecognized role for this intermediate in central carbon metabolism in AML pathophysiology.
α-KGDH–mediated α-KG production is essential for fueling the TCA cycle and maintaining aspartate pools, which are crucial for de novo nucleotide biosynthesis in AML cells. Inhibition of OGDH, the E1 subunit of α-KGDH, disrupts this process, resulting in impaired nucleotide metabolism. This disruption leads to cell cycle arrest, promotes cellular differentiation, and reduces the proliferative capacity of AML cells. The figure was created with BioRender.com.
α-KGDH–mediated α-KG production is essential for fueling the TCA cycle and maintaining aspartate pools, which are crucial for de novo nucleotide biosynthesis in AML cells. Inhibition of OGDH, the E1 subunit of α-KGDH, disrupts this process, resulting in impaired nucleotide metabolism. This disruption leads to cell cycle arrest, promotes cellular differentiation, and reduces the proliferative capacity of AML cells. The figure was created with BioRender.com.
α-KGDH is a key enzyme in the tricarboxylic acid (TCA) cycle, catalyzing the conversion of α-KG to succinyl coenzyme A (succinyl-CoA) in the mitochondria. This multi-subunit complex consists of 3 subunits: OGDH (E1), DLST (E2), and DLD (E3), each with a distinct function. E1 decarboxylates α-KG, E2 transfers the resulting succinyl group to CoA, and E3 regenerates the oxidized form of the lipoic acid cofactor by transferring electrons to nicotinamide adenine dinucleotide (NAD). α-KGDH is central to mitochondrial redox homeostasis by producing NAD plus hydrogen (NADH), which fuels the electron transport chain. It also regulates α-KG flux through the TCA cycle, generating essential intermediates for downstream metabolic pathways. This activity also impacts α-KG–dependent dioxygenases,7 which modulate the epigenetic landscape in cancer cells. In addition, α-KGDH supports amino acid metabolism, protein synthesis, and de novo nucleotide biosynthesis.8
To investigate the role of α-KGDH in AML, the authors used doxycycline-inducible short hairpin RNA to deplete OGDH in murine and patient-derived xenograft models of AML. OGDH depletion impaired leukemic progression and extended survival in models of mixed lineage leukemia (MLL)-fusion and p53-mutant AML. Notably, in relapsed cases, residual AML cells regained OGDH expression, highlighting its critical role in disease progression. Metabolic profiling and isotope tracing revealed that OGDH inhibition disrupted the TCA cycle, reducing intermediates such as fumarate, malate, and aspartate (derived from oxaloacetate). OGDH depletion also reduced NADH production and increased reductive carboxylation9; however, this mechanism could not restore aspartate levels. As a nonessential amino acid required for nucleotide biosynthesis, aspartate depletion impaired de novo purine and pyrimidine synthesis, as evidenced by the accumulation of upstream purine intermediates and depletion of downstream products. Isotope tracing confirmed reduced nucleotide synthesis from glucose and glutamine in OGDH-depleted AML cells. Furthermore, OGDH inhibition altered amino acid metabolism by shifting transaminase activity from production to consumption, reducing serine and alanine levels, which are critical for AML cell viability. These findings underscore OGDH’s multifaceted role in coordinating intracellular metabolism to sustain AML progression.
Interestingly, OGDH inhibition impaired cell cycle progression and induced AML cell differentiation. The MLL-fusion AML model showed increased expression of myeloid differentiation markers and reduced expression of stemness markers, while the p53-mutant AML line exhibited enhanced erythroid differentiation. These phenotypes were accompanied by reduced cell cycle progression but no detectable changes in 5-hydroxymethylcytosine levels. In addition, low expression of the aspartate transporter SLC1A3 in p53-mutant AML cells reduced their ability to import aspartate from extracellular sources. Overexpression of SLC1A3 or aspartate supplementation rescued proliferation and prevented differentiation, further confirming the dependence of AML cells on OGDH-generated aspartate pools. Millman et al also explored the broader implications of OGDH dependency by integrating CRISPR screening data with metabolomic profiles across diverse cancer types. Strong codependencies were observed between OGDH and enzymes involved in de novo nucleotide biosynthesis, but not in salvage pathways.10 This dependency correlated with nucleoside monophosphate levels, rather than population doubling times or OGDH transcript abundance, suggesting that AML's reliance on OGDH is primarily driven by its role in nucleotide biosynthesis.
Collectively, this study uncovers a new functional link between OGDH activity and the aspartate pool for nucleotide biosynthesis in AML and other cancers (see figure). It also highlights the therapeutic potential of targeting OGDH in AML treatment. Importantly, the selective dependency of AML cells on OGDH may provide a therapeutic window. Combination therapies that induce metabolic stress or target complementary pathways, such as nucleotide biosynthesis or aspartate transport, could further enhance the effectiveness of OGDH inhibition.
Despite recent advances in targeting metabolism for hematologic disorders, challenges remain before broader clinical application can be achieved.3 A major hurdle is translating findings from in vitro or ex vivo models to in vivo settings, where the metabolic environment often differs from that of current experimental models. Cancer cells exhibit remarkable adaptability, frequently hijacking alternative metabolic pathways to sustain growth and survival, which increases the risk of therapy resistance. Moreover, the potential toxicity of metabolic inhibitors requires careful assessment, as many targeted metabolic pathways are shared between malignant and healthy cells. Single-agent therapies often produce only transient responses, highlighting the need for combination therapies to achieve durable clinical benefits. Furthermore, identifying reliable predictive biomarkers is crucial for optimizing patient selection and personalizing metabolism-targeted therapies. Overcoming these challenges will pave the way for translating promising preclinical findings into effective, long-lasting treatments for AML and other hematologic malignancies.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal