Abstract
Thrombopoietin (TPO) is the primary hematopoietic growth factor involved in the regulation of platelet production. Although the kidney, liver, bone marrow (BM), and spleen have been identified as the major sources of TPO production, the precise cellular location of TPO mRNA expression in these tissues remains unknown. We have identified the cells expressing TPO mRNA in the human kidney, liver, and BM using an in situ hybridization assay. In the BM of individuals with normal platelet counts, the hybridization signal was too weak to allow identification of the TPO mRNA expressing cells. However, in thrombocytopenic subjects with aplastic anemia, postchemotherapy marrow aplasia, and immune thrombocytopenia, the stromal cells showed strong TPO mRNA expression. In the human subjects with normal platelet counts, the cells of the proximal convoluted tubules of the kidney showed consistent positive staining whereas the signal in the cells of the distal convoluted tubules was less consistent. Strong hybridization signal was also evident in the hepatocytes. The hybridization signal in the spleen, even in thrombocytopenic subjects, was too weak to allow confident identification of the cells expressing TPO mRNA. In all subjects, the interstitial cells and endothelial cells of the liver and spleen, the renal peritubular cells, and the hematopoietic precursor cells of the BM showed no TPO mRNA expression. Our data suggest that TPO mRNA expression in the human BM may be modulated by platelet mass.
THROMBOPOIETIN (TPO) is the major cytokine involved in the growth and development of megakaryocytes and the regulation of platelet production. It was previously believed that megakaryocyte colony-stimulating factor (Meg-CSF ) stimulated the proliferation of progenitor cells of the megakaryocytic lineage whereas TPO controlled megakaryocyte development.1 However, recent data suggest that Meg-CSF and TPO may be the same cytokine which has both megakaryocyte growth and differentiation promoting activities.2
The binding of TPO to its receptor, c-mpl, influences megakaryocyte development both in vitro and in vivo.3-7 The c-mpl proto-oncogene is a member of the cytokine receptor superfamily and was used to identify its ligand, termed thrombopoietin (c-mpl ligand).8
The regulation of TPO expression has become a topic of recent research interest. One mechanism is that TPO plasma levels are dependent on the rate of platelet/megakaryocyte TPO receptor-mediated uptake and destruction. When platelet levels are high, an increased amount of c-mpl ligand (TPO) is taken up by platelets and megakaryocytes resulting in a decrease in circulating levels of the cytokine, thereby limiting megakaryocyte production.9 An alternative mechanism is that TPO gene expression may be regulated by feedback control at the cellular level and TPO mRNA levels may vary accordingly.2 However, these two mechanisms are not mutually exclusive.
Recently reported experimental findings in mice suggest that both mechanisms may be involved in the regulation of platelet production. Stoffel et al10 showed that the incubation of plasma with increasing concentrations of platelets resulted in a dose-dependent depletion of plasma TPO. These investigators together with McCarty et al11 both found that TPO mRNA expression in the liver and kidney were unaffected by changes in circulating platelet levels. These data are consistent with the former mechanism of regulation.
On the other hand, McCarty et al11 showed that TPO mRNA expression in the bone marrow (BM) and spleen increased when the animals were made thrombocytopenic after either irradiation and treatment with carboplatinum or administration with rabbit antiplatelet antiserum. The level of TPO mRNA decreased when thrombocytosis was induced by administration of TPO. These observations suggest that there is regulation of TPO gene expression by a feedback control at least in the BM and spleen. However, the relationships between the circulating platelet mass and TPO gene regulation in the BM, spleen, liver, and kidney has not been studied previously in humans.
To gain a better understanding of the regulation of TPO, the exact site of TPO production may need to be elucidated. The precise cellular origin of TPO is not known, although studies using Northern blot analysis have shown that the major sites of TPO production are in the kidney and liver and, to a lesser extent, in the spleen and BM.12 However, there are a variety of cells present in these organs and the exact cell types that produce TPO have yet to be identified.
One of the aims of our study was to identify the precise cell types expressing TPO mRNA in the kidney, liver, BM, and spleen. This was performed using an in situ hybridization assay with biotin labeled oligonucleotide and cDNA probes.13 14 In addition, we studied the regulation of TPO mRNA expression by examining the TPO gene transcripts in the same four tissues of human subjects with normal, increased, and decreased platelet counts.
MATERIALS AND METHODS
Patient material.Tissue sections were obtained from specimens removed with informed consent during surgery or diagnostic procedures through the Department of Anatomical Pathology at both Prince of Wales and Prince Henry Hospitals. These tissues included kidney, liver, BM trephines, and spleen. The characteristics of the patients from whom the tissues were obtained are described in Table 1.
Clinical Details of Patients From Whom Tissues Were Obtained
| Tissue Type . | Platelet Count and Other Clinical Details . |
|---|---|
| BM trephines | |
| (a) Normal platelet count with normocellular marrow | |
| Platelet count = 230 ± 45 × 109/L (mean ± SD) | |
| Stage 1 or 2 Non-hodgkin's lymphoma (n = 3) | |
| Stomach carcinoma for staging (n = 1) | |
| Splenomegaly due to subacute endocarditis (n = 1) | |
| (b) Thrombocytopenia — hypoplastic marrow | |
| Platelet count = 26 ± 15 × 109/L (mean ± SD) | |
| Severe aplastic anemia (n = 4) | |
| Postchemotherapy BM aplasia (n = 3) | |
| (c) Thrombocytopenia — normocellular marrow with increased megakaryocytes | |
| Platelet count = 33 ± 25 × 109/L (mean ± SD) | |
| ITP (n = 5) | |
| Immune thrombocytopenia secondary to Hodgkin's disease (n = 1) | |
| (d) Thrombocytosis | |
| Platelet count = 993 × 109/L (mean) | |
| Thrombocytosis secondary to an inflammatory myosarcoma of the mesentery (n = 1) | |
| Thrombocytosis due to sepsis (n = 1) | |
| Liver and Spleen | (a) Normal platelet counts with normal liver and spleen functions |
| Platelet count = 232 ± 109 × 109/L (mean ± SD) | |
| Liver resection due to liver metastatic cancer but normal parts of the tissue were used in the study (n = 3) | |
| Spleen removed because of traumatic tear from stab wound (n = 1) | |
| Spleen removed from ITP patient after platelet count had been normalized for 2 wk (n = 1) | |
| (b) Thrombocytopenia | |
| Platelet count = 64 ± 36 × 109/L (mean ± SD) | |
| ITP (n = 2) | |
| Immune thrombocytopenia secondary to Hodgkin's disease (n = 1) | |
| Liver biopsies were performed during splenectomy (n = 2) | |
| Kidney | (a) Normocellular kidney with normal platelet count |
| Platelet count = 289 ± 71 × 109/L (mean ± SD) | |
| Kidneys were removed because of renal cell carcinoma but normal parts of the tissue were used for the study (n = 5) | |
| (b) Thrombocytopenia | |
| Platelet count = 70 × 109/L (mean) | |
| Renal biopsy was performed because of systemic lupus erythematosus (n = 1) and proteinuria (n = 1) | |
| Although some glomeruli were affected by the underlying disease the renal tubules were essentially normal |
| Tissue Type . | Platelet Count and Other Clinical Details . |
|---|---|
| BM trephines | |
| (a) Normal platelet count with normocellular marrow | |
| Platelet count = 230 ± 45 × 109/L (mean ± SD) | |
| Stage 1 or 2 Non-hodgkin's lymphoma (n = 3) | |
| Stomach carcinoma for staging (n = 1) | |
| Splenomegaly due to subacute endocarditis (n = 1) | |
| (b) Thrombocytopenia — hypoplastic marrow | |
| Platelet count = 26 ± 15 × 109/L (mean ± SD) | |
| Severe aplastic anemia (n = 4) | |
| Postchemotherapy BM aplasia (n = 3) | |
| (c) Thrombocytopenia — normocellular marrow with increased megakaryocytes | |
| Platelet count = 33 ± 25 × 109/L (mean ± SD) | |
| ITP (n = 5) | |
| Immune thrombocytopenia secondary to Hodgkin's disease (n = 1) | |
| (d) Thrombocytosis | |
| Platelet count = 993 × 109/L (mean) | |
| Thrombocytosis secondary to an inflammatory myosarcoma of the mesentery (n = 1) | |
| Thrombocytosis due to sepsis (n = 1) | |
| Liver and Spleen | (a) Normal platelet counts with normal liver and spleen functions |
| Platelet count = 232 ± 109 × 109/L (mean ± SD) | |
| Liver resection due to liver metastatic cancer but normal parts of the tissue were used in the study (n = 3) | |
| Spleen removed because of traumatic tear from stab wound (n = 1) | |
| Spleen removed from ITP patient after platelet count had been normalized for 2 wk (n = 1) | |
| (b) Thrombocytopenia | |
| Platelet count = 64 ± 36 × 109/L (mean ± SD) | |
| ITP (n = 2) | |
| Immune thrombocytopenia secondary to Hodgkin's disease (n = 1) | |
| Liver biopsies were performed during splenectomy (n = 2) | |
| Kidney | (a) Normocellular kidney with normal platelet count |
| Platelet count = 289 ± 71 × 109/L (mean ± SD) | |
| Kidneys were removed because of renal cell carcinoma but normal parts of the tissue were used for the study (n = 5) | |
| (b) Thrombocytopenia | |
| Platelet count = 70 × 109/L (mean) | |
| Renal biopsy was performed because of systemic lupus erythematosus (n = 1) and proteinuria (n = 1) | |
| Although some glomeruli were affected by the underlying disease the renal tubules were essentially normal |
Tissues.The sections of kidney were fixed in B5 fixative: 2% (vol/vol) formaldehyde, 6% (vol/vol) mercuric chloride, and 1.25% (vol/vol) sodium acetate. The BM trephines were fixed in 10% (vol/vol) buffered formalin containing 2% (vol/vol) acetic acid (pH 4.0) and decalcified for 24 hours in 14% (wt/vol) sterile EDTA (pH 7.0) before processing, while liver and spleen sections were fixed in 10% (vol/vol) neutral buffered formalin. All sections were processed routinely in an automated tissue processor and embedded in paraffin wax. Tissue sections of 3 to 5 mm were cut and placed on chrome-alum gelatin coated slides. These slides were air-dried in an incubator and the paraffin melted in an oven at 60°C for 30 minutes before dewaxing.
Slides were dewaxed in three changes of xylene through decreasing concentrations of alcohol and two changes of reagent grade water (Continental Water Systems, Sydney, Australia). Many tissues including kidney and liver contain endogenous biotin, which was successfully blocked using a biotin blocking kit (Dakopatts, Glostrup, Denmark), according to the manufacturer's instructions. Slides were then brought back to absolute alcohol before being air-dried in an incubator. A template of parafilm (American Can Co, Greenwich, CT) was cut and placed around the section and melted to the slide by gentle heat on a hot plate13 as previously described.
Probes.A biotin-labeled 28-mer oligonucleotide probe complementary to the mRNA of interest was synthesized with equal (GC):(AT) content (Bresatec, Adelaide, South Australia). The oligonucleotide probe (5′-ACAAGGATCCCAATGCCATCTTCCTGAG3′ ) was designed with the help of published TPO coding sequence.3 Hybridization solution without probe was used to ensure that the endogenous biotin was completely blocked and produced no more signal than background. The sense strand of the oligonucleotide was used as a specific control (negative control). All the oligonucleotides were checked against the Genebank Database to ascertain that sequences were as unique as possible to the target of interest, using the BLAST-Align program,15 located at the internet site http://www.ncbi.nlm.nih.gov/BLAST/
The human TPO plasmid (donated by Dr J. Rasko/Prof D. Metcalf, Walter and Eliza Hall Institute of Medical Research, Victoria, Australia) conferred ampicillin resistance in Escherichia coli. The plasmid was cut by EcoRI (Promega, Madison, WI) and run on a 1% agarose-TBE (Tris/Boric acid/ EDTA) gel to allow identification of the insert by size determination. The cDNA probe was then purified using the Wizard Maxiprep DNA Purification Kit (Promega). The purified plasmid was biotin labeled using Photobiotin (Bresatec) according to the manufacturer's instructions. The plasmid vector pBR322 was used as a negative control. Both β-actin mRNA and alkaline phosphatase mRNA were used as positive controls.
In situ hybridization for TPO mRNA.Hybridization buffer, composed of 3 mL 20× SSC (175.4 g NaCl, 88.2 g sodium citrate in 1 L), 0.25 mL 10 mg/mL herring sperm DNA in 10 mmol/L EDTA (Promega), 0.25 mL 1 mg/mL acetylated bovine serum albumin (BSA) (Promega), 0.5 mL sterile water, and 5 mL formamide (Progen, Queensland, Australia), in a final volume of 9 mL, was mixed well and stored at −20°C until use. Oligonucleotide probe (100 μL probe and 900 μL hybridization buffer), at a final concentration of 100 ng/mL, was then applied to each section and the sections were sealed with a coverslip to ensure even coverage with hybridization solution. They were heated to 65°C in a humidified chamber for 5 minutes to destabilize mRNA secondary structures, and then incubated overnight at 42°C.
In the case of the cDNA probe, 100 μL of photobiotin labeled probe (5 μg) and 20 μL of 10× SSC (sodium chloride/sodium citrate) were placed in a sterile Eppendorf tube and heated to 90°C for 5 minutes to dissociate the double-stranded DNA. This was mixed well, chilled on ice, and then 900 μL of hybridization buffer was added. The hybridization buffer, composed of 12.5 mL 20× SSC, 5 mL 2.5 mg/mL Herring sperm DNA in 100 mmol/L EDTA (Promega), 5 g Dextran sulfate MW 500 000 (Sigma, St Louis, MO), 25 mL formamide (Progen), and sterile water to a final volume of 45 mL, was prepared beforehand and stored at −20°C. To each section, 100 μL of the appropriate cDNA probe at a final concentration of 5 μg/mL was applied. The sections were then sealed with a coverslip and placed in a humidified chamber at 42°C overnight.
Posthybridization and detection.The posthybridization and detection conditions varied between the oligonucleotide probes and the longer cDNA probes. After overnight hybridization with the oligonucleotide probes, coverslips were removed in 2× SSC-0.1% (wt/vol) sodium dodecyl sulfate (SDS). The hybridization buffer was decanted from the slides and any nonspecific sequences were removed by washing in 2× SSC-50% (vol/vol) formamide for 10 minutes at 42°C. This was followed by two washes in 2× SSC-0.1% (wt/vol) SDS for 10 minutes at 42°C. The slides were then washed four times with 2× SSC at room temperature to remove the SDS.
The slides incubated with the cDNA probe were washed in 2× SSC-50% (vol/vol) formamide for 15 minutes after overnight hybridization to remove any nonspecific sequences. This was followed by washing two sequential applications of 2× SSC-0.1% (wt/vol) SDS and 0.4× SSC-0.1% (wt/vol) SDS at 42°C for 15 minutes each. The slides were then washed with 0.4× SSC to remove the SDS. All slides were placed in TBS Buffer 1 (0.1 mol/L Tris, 0.5 mol/L NaCl, 5 mmol/L MgCl2 , 0.025% [vol/vol] Triton X-100, 0.025% [wt/vol] BSA [Sigma], pH 7.6) for 5 minutes before the detection of the biotin label.
Biotin on the hybridized probes was detected using a streptavidin and biotinylated alkaline phosphatase–based in situ hybridization detection system (Dakopatts) with slight modification of the manufacturer's instructions. The following reagents were applied sequentially for 10 minutes each with 4 TBS Buffer 1 washes between applications: (1) streptavidin 1:100 dilution, (2) biotinylated alkaline phosphatase 1:100 dilution, (3) streptavidin 1:500 dilution, (4) biotinylated alkaline phosphatase 1:100 dilution, with steps (3) and (4) repeated a further three times for the oligonucleotide probes.
Visualization of detection systems.After the detection procedures, alkaline phosphatase activity was demonstrated using the Fast-Red naphthol-capture method. Substrate development was usually completed within 30 minutes at which time the coverslips were removed in TBS Buffer 2 (0.1 mol/L Tris, 0.1 mol/L NaCl, 10 mmol/L MgCl2 , pH 9.2). The sections were rinsed in three changes of distilled water before counterstaining progressively with Mayer's haematoxylin for 15 to 20 minutes. The slides were washed with tap water, blued with weak ammonia solution (1 drop concentrated ammonia in 200 mL water) for 30 seconds, washed again in water, and then the parafilm templates removed and sections mounted with glycerol-gelatin (Sigma).
With the use of oligonucleotide probes, the coverslips were removed in Buffer 3 (0.1 mol/L Tris, 0.5 mol/L NaCl, 10 mmol/L MgCl2 , pH 9.2). The high salt concentration stabilized the oligonucleotide probes, preventing them from being removed during washing.
RESULTS
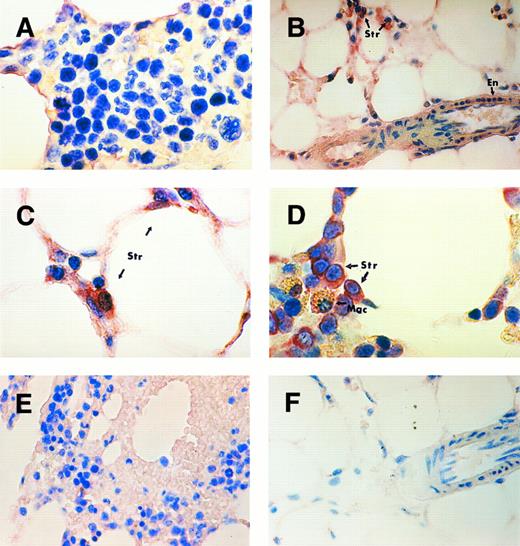
BM.In this study, a direct strategy was used to identify the type of cells which express TPO mRNA in the kidney, liver, spleen, and BM. The probes designed for the in situ hybridization procedure contained sequences complementary to the mRNA of interest. In patients with normal platelet counts, the hybridization signal observed in the BM was too weak to allow confident identification of the cells expressing TPO mRNA (Fig 1A). However, in patients with thrombocytopenia the stromal cells of the marrow showed a definite increase in hybridization signal compared to marrow sections from patients with normal platelet counts. There was a clear difference in the signal development in the stromal cells between subjects with thrombocytopenia from marrow aplasia (aplastic anemia and postchemotherapy aplasia) (Fig 1B and C) and subjects with thrombocytopenia due to increased peripheral platelet clearance (idiopathic thrombocytopenic purpura [ITP] or secondary immune thrombocytopenia, Fig 1D). The signal development in the trephine section of the subject with Hodgkin's disease and secondary immune thrombocytopenia (Fig 1D) was the strongest with an increase in the staining intensity in some endothelial cells. However, the myeloid and erythroid precursors, megakaryocytes, and macrophages showed negative staining (Fig 1C).
TPO mRNA expression assessed by in situ hybridization on paraffin-embedded human BM trephine sections. BM of a subject with normal platelet count (A) showing weak signal, perhaps in stromal cells but negative staining in hematopoietic precursor cells. BM trephine of a patient with severe aplastic anemia (B and C) and a patient with ITP (D) showing strong hybridization signal in stromal cells (Str) and weak signal in endothelial cells (En). The macrophages (Mac) with golden brown hemosiderin granules show negative staining (D). BM of a subject with secondary thrombocytosis (E) showing no visible difference in staining intensity compared to that of the subject with a normal platelet count. Trephine section of aplastic anemia patient (F ) probed with plasmid vector pBR322 as a negative control. Original magnifications: (B, E, and F ) ×400; (A, C, and D) ×1,000.
TPO mRNA expression assessed by in situ hybridization on paraffin-embedded human BM trephine sections. BM of a subject with normal platelet count (A) showing weak signal, perhaps in stromal cells but negative staining in hematopoietic precursor cells. BM trephine of a patient with severe aplastic anemia (B and C) and a patient with ITP (D) showing strong hybridization signal in stromal cells (Str) and weak signal in endothelial cells (En). The macrophages (Mac) with golden brown hemosiderin granules show negative staining (D). BM of a subject with secondary thrombocytosis (E) showing no visible difference in staining intensity compared to that of the subject with a normal platelet count. Trephine section of aplastic anemia patient (F ) probed with plasmid vector pBR322 as a negative control. Original magnifications: (B, E, and F ) ×400; (A, C, and D) ×1,000.
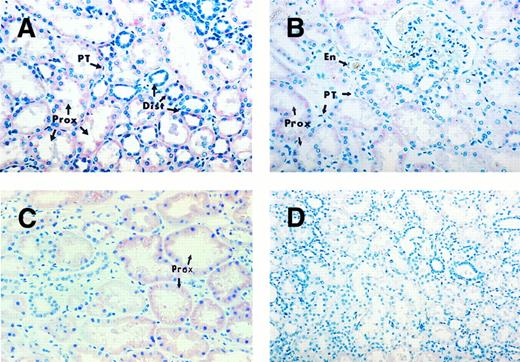
Human kidney sections exhibiting TPO mRNA expression. Kidney sections of individuals with normal platelet counts, probed with TPO cDNA probe (A), TPO oligonucleotide probe (B), and the sense strand of the TPO mRNA as a negative control (D). Kidney section of a thrombocytopenic patient probed with TPO cDNA probe (C). Strong hybridization signal in proximal convoluted tubules (Prox) and less consistent staining in the distal convoluted tubules (Dist). Peritubular cells (PT) and endothelial cells (En) remain negative. Original magnifications: (A, B, and C) ×396; (D) ×240.
Human kidney sections exhibiting TPO mRNA expression. Kidney sections of individuals with normal platelet counts, probed with TPO cDNA probe (A), TPO oligonucleotide probe (B), and the sense strand of the TPO mRNA as a negative control (D). Kidney section of a thrombocytopenic patient probed with TPO cDNA probe (C). Strong hybridization signal in proximal convoluted tubules (Prox) and less consistent staining in the distal convoluted tubules (Dist). Peritubular cells (PT) and endothelial cells (En) remain negative. Original magnifications: (A, B, and C) ×396; (D) ×240.
In patients with reactive thrombocytosis, the hybridization signal in the stromal cells and other cells in the marrow was not significantly different from that in the patients with normal platelet counts (Fig 1E). It is possible that the in situ hybridization assay was unable to detect any decrease in signal in thrombocytotic individuals from the very weak staining in the normal controls. Specificity of the hybridization signal was confirmed by the absence of signal in the trephine section probed with the plasmid vector pBR322 as a negative control (Fig 1F ).
Kidney, liver, and spleen.In the kidney sections from patients with normal platelet counts probed with the TPO cDNA probe, the cells of the proximal convoluted tubules showed consistent positive staining, whereas the cells of the distal convoluted tubules showed more sporadic staining (Fig 2A). The kidney sections probed with the oligonucleotide probe (Fig 2B) showed similar hybridization patterns, but was less intense than that obtained with the former. As a result of the increased signal strength with the cDNA probe, the use of the oligonucleotide probe was discontinued. The hybridization signal in the kidney sections of thrombocytopenic individuals was essentially the same as that of the subjects with normal platelet counts (Fig 2C). Interstitial cells, specifically the renal peritubular cells and endothelial cells, showed negative staining (Fig 2B). Specificity of the hybridization signal was confirmed by the negative result using the sense strand of TPO mRNA as a negative control (Fig 2D).
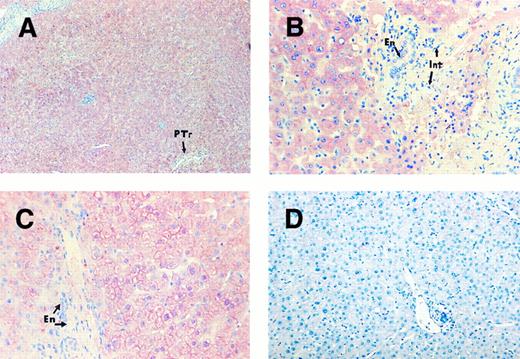
Moderate to strong cytoplasmic staining was observed in the hepatocytes in subjects with normal platelet counts. There was a suggestion of a patchy distribution with somewhat stronger signal development around the peri-portal region of the liver (Fig 3A), whereas the endothelial cells remained unlabeled (Fig 3B). There was no visible increase in the staining intensity in the liver sections of thrombocytopenic individuals (Fig 3C) compared with that of the subjects with normal platelet counts. Control sections probed with the plasmid vector pBR322 showed absence of signal (Fig 3D).
TPO mRNA expression assessed by in situ hybridization on human liver sections. Liver sections of a patient with normal platelet counts (A and B) and a patient with ITP (C), probed with TPO cDNA probe. Strong signal development in hepatocytes but negative staining in endothelial cells (En) and interstitial cells (Int). Patchy signal distribution around portal tract (PTr). Liver section (D) from an individual with normal platelet count probed with plasmid vector pBR322 as a negative control. Original magnification: (A) ×108; (B and C) ×396; (D) ×240.
TPO mRNA expression assessed by in situ hybridization on human liver sections. Liver sections of a patient with normal platelet counts (A and B) and a patient with ITP (C), probed with TPO cDNA probe. Strong signal development in hepatocytes but negative staining in endothelial cells (En) and interstitial cells (Int). Patchy signal distribution around portal tract (PTr). Liver section (D) from an individual with normal platelet count probed with plasmid vector pBR322 as a negative control. Original magnification: (A) ×108; (B and C) ×396; (D) ×240.
Human spleen sections probed with TPO cDNA probe. Splenic sections of an individual with normal platelet count (A) and an individual with ITP (B and C). Faint, almost negative hybridization signal in the splenic cells, but clearly negative staining in the endothelial cells (En) and fibroblasts (Fb). Spleen section from patient with normal platelet count (D) probed with plasmid vector pBR322 as a negative control. The orange staining cells are erythroid cells (Ery) that do not show any positive signal development. Original magnification: (A and C) ×400; (B and D) ×240.
Human spleen sections probed with TPO cDNA probe. Splenic sections of an individual with normal platelet count (A) and an individual with ITP (B and C). Faint, almost negative hybridization signal in the splenic cells, but clearly negative staining in the endothelial cells (En) and fibroblasts (Fb). Spleen section from patient with normal platelet count (D) probed with plasmid vector pBR322 as a negative control. The orange staining cells are erythroid cells (Ery) that do not show any positive signal development. Original magnification: (A and C) ×400; (B and D) ×240.
The hybridization signal in the splenic tissue of patients with normal platelet counts was too weak to precisely identify the cells expressing TPO mRNA (Fig 4A). There was no increase in the signal intensity, even in the spleen of thrombocytopenic patients (Fig 4B and C). The interstitial cells and endothelial cells remained negative. Once again, spleen sections probed with plasmid vector pBR322 showed absence of signal (Fig 4D).
In addition, no specific pattern of hybridization was shown in other tissues tested, including brain, lung, stomach, and thyroid.
DISCUSSION
In this study, we found strong TPO mRNA expression in the hepatocytes in the human liver and the convoluted tubular cells in the human kidney. The expression of TPO mRNA was somewhat stronger in the peri-portal region of the liver. In the kidney, the hybridization signal was consistently strong in the proximal tubular cells but was more variable in the distal tubular cells. In the BM, it was the stromal cells that expressed TPO mRNA. However, this became obvious only when TPO mRNA expression was upregulated in the thrombocytopenic subjects. In the individuals with normal platelet counts, the strength of the hybridization signal was too weak to allow confident identification of the cells that expressed TPO mRNA. We were unable to localize the cells expressing TPO mRNA in the spleen because the TPO gene expression remained weak in individuals with normal and reduced platelet counts. Our data are consistent with the results of previous studies which showed TPO mRNA expression in murine and human renal and liver tissues and the human hepatocellular carcinoma cell line Hep-G2, and TPO production by isolated rat liver cells and cell lines (H4-II-E, McA-RH8994, and HTC).11,16,17 In contrast to previous studies that used Northern blot analysis, RNase protection, and reverse transcription-polymerase chain reaction (RT-PCR),11,16,18,19 the present study uses a highly sensitive in situ hybridization technique13 14 with biotin-labeled cDNA and oligonucleotide probes that can detect intracellular mRNA of interest without having to extract RNA and bind it to membranes. This in situ technique allows the identification of the precise cell types in the kidney, liver, and BM which express TPO, and the location of TPO expressing cells in relation to other cells and structures in these organs. This information may give useful insights into the micro-environment of these tissues in which TPO is produced.
Our finding that different cell types express TPO mRNA in different tissues is of interest, particularly when some investigators have raised the possibility that TPO production in various tissues may come from the same source, such as endothelial cells, fibroblasts, or tissue macrophages11 following recent reports of cultured fibroblasts and endothelial cells expressing TPO mRNA.16 Our results indicate that this is probably not the case. We did not detect TPO gene transcript in the endothelial cells and interstitial cells in the liver, kidney, and spleen. However, in the BM of thrombocytopenic subjects we found strong TPO mRNA expression in the stromal cells but very weak and variable expression in the endothelial cells. No convincing expression was detected in the macrophages.
Our results may provide further insights into TPO gene regulation. Two possible mechanisms have been postulated for the control of plasma TPO levels.2 Kuter and Rosenberg9 have proposed that plasma TPO concentrations are dependent on circulating platelet mass. Raised platelet levels would lead to increased binding of the cytokine to platelet receptors and consequently increased catabolism, resulting in a lower plasma concentration. On the other hand, lower platelet levels would lead to decreased uptake and catabolism and, hence, a higher plasma TPO concentration. The recently reported phenotype of the NF-E2 knockout mouse is not consistent with this being the sole mechanism of TPO regulation. The platelet counts in the NF-E2−/− animals were markedly reduced and yet the serum TPO levels remained normal.20 The findings of McCarty et al11 and our results showing upregulation of TPO mRNA in the BM in thrombocytopenic mice and human subjects, respectively, would suggest that there is an additional or alternative mechanism.
In the alternative mechanism, the TPO plasma concentrations are probably regulated by feedback control at the level of gene expression. High platelet counts may inhibit TPO gene transcription or decrease stability of TPO gene transcript, possibly via platelet secreted proteins such as platelet factor 4.21 Thrombocytopenia would have the opposite effect. Our data in humans and the findings of Stoffel et al10 and McCarty et al11 in mice would suggest that the feedback regulation occurs predominantly in the BM and probably not in the liver and kidney, as these investigators did not find any increase in TPO mRNA expression in the liver and kidney of thrombocytopenic mice10,11 or human subjects. However, our results suggest that there may be a slight difference in TPO gene regulation between mice and humans in that, unlike the mice, the human spleen may not be involved in the feedback control. We found no evidence of upregulation of TPO mRNA expression in the spleen of human thrombocytopenic subjects. In contrast, the study by McCarty et al11 showed an increase in TPO gene transcript in the splenic tissue of mice rendered thrombocytopenic by either irradiation and carboplatinum or administration of rabbit antimouse platelet sera. These investigators believe that the species difference may be due to the greater role of the spleen as a site of hematopoiesis in the mouse. Even though our results seem reasonably convincing, they were obtained using a nonquantitative assay (in situ hybridization). Confirmation of the data using more quantitative assays are required in the future.
However, the two mechanisms of gene regulation may not be mutually exclusive. The two, and perhaps other mechanisms, may operate in concert. Furthermore, platelets may not be the only cells that catabolize TPO. The findings of a recent study22 suggest that the major determinant of plasma TPO levels may be the megakaryocyte mass rather than the platelet number. In this study, patients with aplastic anaemia had high TPO levels whereas those with ITP had much lower levels, even though both patient groups had severe thrombocytopenia. This finding suggests that megakaryocytes may be more efficient than platelets in TPO uptake and catabolism. The increased megakaryocytes in the BM of patients with ITP may absorb the locally produced TPO preventing any significant spillage into the peripheral blood. However, there may be other explanations for these observations. Further studies are clearly required before we can fully understand all the complexities of TPO regulation.
ACKNOWLEDGMENT
We are extremely grateful for the help provided by the following people: Claire and Brian Walsh (Department of Anatomical Pathology, Prince of Wales Hospital) for the cutting of the tissue sections; Drs Roger Crouch (Department of Anatomical Pathology, Prince of Wales Hospital) and Jim Yong (Department of Anatomical Pathology, Prince Henry Hospital) for the interpretation of the histology; Janette Burgess (Haematology Research, Prince of Wales Hospital) for assistance in the cloning of the plasmids; and Medical Illustrations Unit (Prince of Wales Hospital) for the color photography.
Supported in part by a grant from the National Health and Medical Research Council of Australia.
Address reprint requests to B.H. Chong, MD, Department of Haematology, Prince of Wales Hospital, High St, Randwick, NSW, 2031 Australia.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal