Abstract
Mantle cell lymphoma (MCL) is molecularly characterized by bcl-1 rearrangement and cyclin D1 gene overexpression. Some aggressive variants of MCL have been described with blastic or large cell morphology, higher proliferative activity, and shorter survival. The cyclin-dependent kinase inhibitors (CDKIs) p21Waf1 and p16INK4a have been suggested as candidates for tumor-suppressor genes. To determine the role of p21Waf1 and p16INK4a gene alterations in MCLs, we examined the expression, deletions, and mutations of these genes in a series of 24 MCLs, 18 typical, and 6 aggressive variants. Loss of expression and/or deletions of p21Waf1 and p16INK4a genes were detected in 4 (67%) aggressive MCLs but in none of the typical variants. Two aggressive MCLs showed a loss of p16INK4a expression. These cases showed homozygous deletions of p16INK4a gene by Southern blot analysis. An additional aggressive MCL in which expression could not be examined showed a hemizygous 9p12 deletion. Loss of p21Waf1 expression at both protein and mRNA levels was detected in an additional aggressive MCL. No p21Waf1 gene deletions or mutations were found in this case. The p21Waf1 expression in MCLs was independent of p53 mutations. The two cases with p53 mutations showed p21Waf1 and p16INK4a expression whereas the 4 aggressive MCLs with p16INK4a and p21Waf1 gene alterations had a wild-type p53. p21Waf1 and p16INK4a were expressed at mRNA and protein levels in all typical MCLs examined. No gene deletions or point mutations were found in typical variants. Two typical MCLs showed an anomalous single-stranded conformation polymorphism corresponding to the known polymorphisms at codon 148 of p16INK4a gene and codon 31 of p21Waf1 gene. These findings indicate that p21Waf1 and p16INK4a alterations are rare in typical MCLs but the loss of p21Waf1 and p16INK4a expression, and deletions of p16INK4a gene are associated with aggressive variants of MCLs, and they occur in a subset of tumors with a wild-type p53 gene.
MANTLE CELL lymphoma (MCL) is a malignant lymphoproliferative disorder genetically characterized by chromosomal translocations involving the 11q13 region and its molecular counterpart the bcl-1 rearrangement.1-4 This translocation activates the cyclin D1 gene located 110 to 130 kb downstream from the major breakpoint of this rearrangement.5-7 Cyclin D1 overexpression occurs in virtually all MCL independently of the detection of t(11; 14) translocation or bcl-1 rearrangements, and this overexpression is highly specific of this type of lymphoma.8-11 Cyclin D1 is a G1 cyclin that participates in the control of the cell-cycle progression at the G1 to S phase transition. Cyclin D1 binds to Retinoblastoma protein (pRb) and, coupled with cyclin-dependent kinases (CDKs), is capable of its phosphorylation contributing to the inactivation of the pRb growth suppressive effect.12,13 Different studies have also shown that cyclin D1 may function as an oncogene cooperating with other oncogenes in cellular transformation.14 15 However, the tumorigenic and transforming properties of cyclin D1 seem to be less effective than the conventional oncogenes. The possible additional oncogenic factors cooperating with cyclin D1 in the pathogenesis of MCLs are unknown.
In addition to cyclin D1, other cell-cycle regulatory molecules may participate in the development and progression of human tumors. Particularly, an emerging group of small proteins called cyclin-dependent kinase inhibitors (CDKIs) are considered to be implicated in tumorigenesis as possible tumor-suppressor genes.16,17 These proteins act as negative regulatory elements of cell proliferation by inhibiting the kinase activity of the cyclin/CDK complexes. Among all these molecules, p21Waf1 and p16INK4a are strong candidates to participate in tumor progression. p21Waf1 is a gene induced by wild-type p53 but not mutant p53 and it is implicated in the mechanisms of cell arrest that allow the cell for DNA repair.18,19 p21Waf1 is also involved in mechanisms of cell differentiation and senescence.20 The potential role for p21Waf1 in tumorigenesis has been postulated on the basis of its transcriptional control by p53. In addition, overexpression of p21Waf1 may suppress tumor growth in different experimental models.21,22 However, p21Waf1 point mutations seem to be very rare in human tumors.23 p16INK4a and p15INK4b genes are located on 9p21, a chromosomal region frequently deleted in many human tumors. p16INK4a gene has been found to be deleted, mutated, or downregulated by hypermethylation at high frequency in different types of tumors.16,17,24,25 In hematologic disorders, homozygous deletions are frequently found in acute lymphoblastic leukemias but point mutations seem to be rare.17 The spontaneous development of B-cell lymphomas in the INK4a knock-out mice strongly supports the role of this gene in the pathogenesis of lymphoproliferative disorders.26
The possible alterations of p21Waf1 and p16INK4a in MCLs have not been previously examined. We and others have recently shown that p53 mutations are associated with aggressive variants of MCLs.27-29 Whether p53 mutations may downregulate the transcription rate of p21Waf1 expression in these aggressive MCLs is not known. On the other hand, it has been suggested that p16INK4a, cyclin D1, and pRb are interlinked in the same cell-cycle regulatory pathway.30-33 Oncogenic alterations of p16INK4a and cyclin D1 cooperate synergistically to deregulate cell proliferation in cell lines with normal pRb function.33 The constant cyclin D1 overexpression8 and the presence of an apparently functional pRb34 35 in MCLs make these tumors an interesting in vivo model to analyze the possible cooperating effects of p16INK4a and p21Waf1 in the pathogenesis of these lymphomas.
To determine the possible role of p21Waf1 and p16INK4a gene alterations in the development and progression of MCLs, we have examined their gene structure and expression at both mRNA and protein levels in a series of MCLs previously characterized for cyclin D1 overexpression,8 retinoblastoma status,34 and p53 gene mutations.28 Our results indicate that p21Waf1 and p16INK4a alterations are rare in typical MCLs but the loss of p21Waf1 and p16INK4a expression and deletions of p16INK4a gene are associated with aggressive variants of MCLs, and they occur in a subset of tumors with wild-type p53 gene.
MATERIALS AND METHODS
Case selection.Tumor specimens from 24 MCLs were obtained from the files of the Pathology Department of the Hospital Clinic of Barcelona. The specimens included 18 lymph nodes, 4 spleens, 1 tonsil, and 1 skin biopsy. Eighteen cases were classified as typical MCLs36,37 and 6 cases as aggressive variants of MCLs. These aggressive variants were further classified as blastic MCLs (4 cases) and large cell (“anaplastic”) type of MCL (2 cases) defined according to criteria previously described.38-40 Immunophenotype was analyzed in all cases using immunohistochemistry on frozen tissue sections and/or cell suspensions by flow cytometry. These studies included Ig light and heavy chains, several of the B-cell, and T-cell markers, CD10 and CD23. bcl-1 rearrangement at the major translocation cluster (MTC) locus was detected by Southern blot and/or polymerase chain reaction (PCR) analysis in 6 typical and 2 aggressive cases using previously described techniques.41 Cyclin D1 overexpression was detected in all cases by Northern and/or Western blot analysis.8,42 All of these cases had also been examined for retinoblastoma protein expression34 and p53 gene mutations28 in previous studies. Analysis of p53 mutations of exon 5 to 9 had been examined by single-stranded conformation polymorphism (SSCP), denaturing gradient gel electrophoresis, and direct sequencing as described.28 Cytogenetic analysis was performed in 5 patients (4 typical and 1 aggressive MCL variants) with leukemic expression.34 The clinical and pathologic characteristics of the patients including the proliferative activity of the tumors and the follow-up of the patients have been previously reported.28 34 The mean Ki-67 proliferative index in aggressive variants of MCL was significantly higher (mean, 38.2%; SD, 5.6) than in the typical MCL (mean, 10.3; SD, 5.6) (P < .002). The median overall survival of this series of MCL patients was 49.8 months (range, 2 to 82 months). Patients with aggressive variants had an overall survival (median, 18.3; n = 6) significantly shorter than the patients with typical MCL (median, 49.8, n = 18) (P < .001).
Southern and Northern blot analyses.Genomic DNA was extracted from frozen material in 23 (18 typical and 5 blastic) cases using Proteinase K/RNAse treatment and phenol-chloroform extraction. DNA from each case (10 μg) was digested with EcoRI, HindIII, and BamHI, separated on 0.8% agarose gels, and transferred to Hybond-N membranes (Amersham, Buckinghamshire, UK). The membranes were prehybridized, hybridized with the p21Waf1 and p16INK4a probes, and washed as previously described.8 The bcl-1 MTC probe was also used in all Southern blots. To evaluate the intensity of the signals of the Southern blot analysis, the autoradiographic signals were quantified using a UVP-5000 video densitometer (UVP, San Gabriel, CA). Intensities of p16INK4a and p21Waf1 bands were normalized to the bcl-1 MTC control bands.
Total RNA was isolated from frozen tissues in 17 cases (12 typical and 5 aggressive variants) by guanidine isothiocyanate extraction and cesium chloride gradient centrifugation.8 Eight (for p21Waf1 analysis) to 20 (for p16INK4a analysis) micrograms of total RNA were electrophoresed on a denaturing 1.2% agarose formaldehyde gel and transferred to Hybond-N membranes (Amersham). The membranes were prehybridized, hybridized with the p21Waf1 and p16INK4a probes, and washed as previously described.8
Probes.Probes were radiolabeled using a random primer DNA labeling kit (Promega Corp, Madison, WI) with [α-32P]dCTP. Human p21Waf1 cDNA probe was obtained by reverse transcriptase-PCR (RT-PCR) from poly A RNA and cloned using standard techniques. For PCR amplification, the following primers were used: 5′-GCGCCATGTCAGAACCGGCTG-3′ and 5′-GCAGGCTTCCTGTGGGCGGAT-3′. The PCR conditions were 95°C for 1 minute, 60°C for 30 seconds, 75°C for 2 minutes, and a total number of 30 cycles. The identity of the amplified fragment was confirmed by sequencing. The p16INK4a probes used were a 0.8-kb EcoRI-Xho I fragment of the p16INK4a cDNA clone kindly provided by Dr M. Serrano,43 and a fragment of exon 2 obtained by PCR using primers previously described.44 The identity of the amplified fragment was confirmed by sequencing.
SSCP analysis.SSCP analysis was used to screen for p16INK4a and p21Waf1 gene mutations according to a modified protocol of a previous described method.45 Exons 1 and 2 of p16INK4a gene were amplified by PCR using a simple set of flanking intronic primers. Primers for exon 1 were 5′- GAAGAAAGAGGAGGGGCTG-3′ and 5′- GCGCTACCTGATCCAATTC-3′, and primers for exon 2 were 5′- CTCTACACAAGCTTCCTTTCC-3′ and 5′- GGGCTGAACTTTCTGTGCTGG-3′. Amplifications were performed with 0.2 μg of genomic DNA, 2.5 U of Taq polymerase (GIBCO-BRL, Gaithersburg, MD), 0.5 mmol/L of each primer, 100 μmol/L of dNTPs, 5% dimethyl sulfoxide, and PCR buffer in a final volume of 50 μL. We used a “touch-down” PCR strategy for the amplification of both exons. Conditions were 1 cycle at 95°C for 5 minutes, 4 cycles at 94°C for 45 seconds, at 68°C for 1 minute, and at 72°C for 1 minute; 4 cycles with annealing temperature at 64°C; 4 cycles with annealing temperature at 62°C; and 30 cycles with annealing temperature at 60°C. Previously to SSCP analysis, the PCR product of both exons were digested with Sma I.
Exon 2 of p21Waf1 gene contains 90% of the coding sequence. This exon was amplified in two fragments using primers 5′-CATAGTGTCTAATCTCCGCCGT-3′ and 5′-GCCTGCCTCCTCCCAACTCATC-3′ for the 5′ segment of exon 2 (fragment 2a), and 5′-TGCCCAAGCTCTACCTTCCCAC-3′ and 5′-AGCCCTTGGACCATGGATTCTG-3′ for the 3′ segment of exon 2 (fragment 2b) as previously described.23 The PCR mixture contained 0.2 μg of genomic DNA, 2.5 U of Taq, 0.5 μmol/L each primer, 100 mmol/L dNTPs, and PCR buffer in a final volume of 50 μL. The reaction was performed for 30 cycles at 94°C for 40 seconds, at 59°C for 30 seconds, and at 72°C for 2 minutes.23
The p16INK4a and p21Waf1 amplified products were diluted sixfold in formamide-dye loading buffer, incubated for 3 minutes at 95°C, and immediately cooled on ice; 20 μL was loaded on a 15% nondenaturing polyacrylamide gel with or without 10% glycerol for p16INK4a, and on a 10% nondenaturing polyacrylamide gel with or without 7% glycerol for p21Waf1 samples. p16INK4a samples were electrophoresed at 150 V for 14 hours at room temperature, and p21Waf1 samples at 150 V for 16 hours at room temperature. The gels were developed using a silver staining procedure previously described.46
DNA sequencing.p16INK4a and p21Waf1 genes were sequenced using a commercial cycle sequencing kit (Perkin Elmer, Branchburg, NY) and 33P dATP. A total of 0.5 μL of the p16INK4a gene PCR products was used as template for sequencing. The primers described above and two internal primers for exon 2, 5′-ACTCTCACCCGACCCGTGCA-3′ and 5′-AGCTCCTCAGCCAGGTCCA-3′ were used for the sequencing reaction at a final concentration of 0.5 μmol/L. The reaction was performed according to the instructions supplied by the manufacturer. The reaction was performed for 35 cycles at 94°C for 30 seconds, 60°C for 30 seconds, and 70°C for 30 seconds.
The two coding exons of p21Waf1 gene were sequenced similarly. Exon 2 was amplified with oligonucleotides 5′- GCGCCATGTCAGAACCGGC-3′ and 5′- GAGAATCCTGGTCCCTTAC-3′, and exon 3 was amplified with oligonucleotides 5′-GCCCCCCACTGTCTTCCT-3′ and 5′- GCGCTTCCAGGACTGCAGG-3′. PCR conditions for amplification consisted of a 10-minute denaturalization step at 94°C (without adding the enzyme), followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C. The same primers were used for the sequencing reaction.
The final amplified products were diluted twofold in formamide-dye loading buffer. Samples were denatured for 3 minutes at 95°C and 2 μL was analyzed in a denaturing 6% polyacrylamide/8 mol/L urea sequencing gel for 2 or 3 hours at 55 W. The gels were dried under vacuum at 85°C and exposed to an x-ray film at room temperature for 3 days without intensifier screen. The presence of a mutation was confirmed by sequencing the other DNA strand.
Western blot analysis.Protein extraction was obtained from additional frozen tissue available in 4 aggressive MCLs and 11 typical MCLs. Protein samples were also extracted from HeLa and Jurkatt cell lines to be used as positive and negative controls. In each case, 10 frozen sections of 30 μm were incubated in 300 μL of ice-cold lysis buffer (50 mmol/L Tris-Cl, pH 8, 150 mmol/L NaCl, 0.4 mmol/L EDTA, 10 mmol/L NaF, 0.02% sodium azide, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, and 0.5% sodium deoxycholate) containing 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 1 μg/mL α-1-antitrypsine for 20 minutes at 4°C. The cell debris was sedimented by centrifugation at 14,000 rpm at 4°C for 25 minutes. The clarified supernatants were collected, and the protein content of the lysate was determined by the Lowry protein assay (Bio-Rad, Hercules, CA). Fifty micrograms of total cellular protein was run per lane on a 15% SDS-polyacrylamide gel and electroblotted to a nitrocellulose membrane (Amersham). Membrane was blocked by overnight incubation in 5% dry milk and 0.1% Tween-20 at 4°C. The blocked membrane was then incubated with the monoclonal antibodies anti-p16INK4a, Clone G175-405 (Pharmingen, San Diego, CA), and anti-p21Waf1 clone EA10 (Oncogene Science, Cambridge, NY) at a final concentration of 1 μg/mL for 1 hour and 30 minutes. The membrane was then washed with phosphate-buffered saline (PBS) 0.1% Tween 20 and exposed to sheep antimouse conjugated to horseradish peroxidase (Amersham) at a 1:1,000 dilution for 1 hour and 30 minutes. After washing, antibody binding was detected by chemiluminescence a detection procedures according to the manufacturer's recommendations (ECL; Amersham).
Immunohistochemical analysis.p21Waf1 protein expression was immunohistochemically assessed in all cases on formalin fixed-paraffin embedded using the-anti-p21Waf1 clone EA10 monoclonal antibody (Oncogene Science). Before the application of the primary antibody an antigen retrieval technique was performed. The deparaffinized and rehydrated slides were placed in 10 mmol/L citrate buffer, pH 6, and heated in the microwave oven for 15 minutes at 700 W. The anti-p21Waf1 antibody was incubated overnight at 4°C. The immunoreaction was detected by means of the streptavidin-biotin-alkaline phosphatase (Biogenex, San Ramon, CA) technique using Fast-Red (Biogenex) as chromogen and levamisole to inhibit endogenous alkaline phosphatase. The slides were counterstained with hematoxylin. The quantification of positive cells was performed using a computerized digital analyzer (Microm Image Processing IMCO; Kontron Electronik, Munich, Germany) to score the nuclei of the tumor cells for the presence of p21Waf1 proteins. The value measured was the percentage of the immunohistochemical stained area against that of the total nuclear area. Nuclear boundary optical density and antibody threshold were adjusted for each case examined. Five or more fields of each tumor were analyzed, until a minimum of 1,000 cells had been examined. The fields were selected in each slide from the areas with greatest number of stained cells.
RESULTS
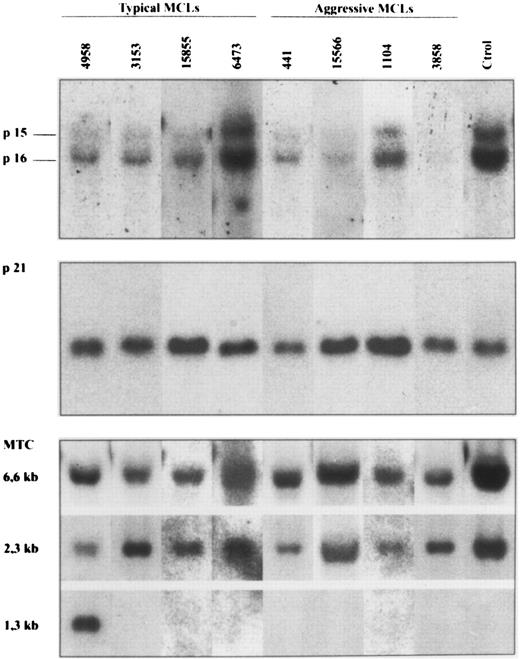
p16INK4a gene analysis.p16INK4a gene was examined by Southern blot and SSCP analysis in a series of 18 typical MCLs and 5 aggressive variants in which genomic DNA was available. The results are summarized in Tables 1 and 2, and in Fig 1. Southern blot analysis showed deletions of p16INK4a gene in two aggressive MCLs with a blastic morphology (cases 3858 and 15566) (Fig 1). These deletions were judged to be homozygous because the DNA signal in these cases was less than 80% of the placental DNA sample. The slight signal observed in case 15566 could be caused by contamination from normal cells. In fact, this sample was a skin biopsy with normal epidermis and stromal tissues. Deletions of the p16INK4a gene in these two cases were also associated with homozygous deletions of the p15INK4b gene (Fig 1). No deletions were found in any of the typical MCLs. In one additional aggressive MCL (case 3747) in which no DNA was available, the cytogenetic analysis showed a complex karyotype with a hemizygous deletion of the short arm of chromosome 9 -del(9)(p12)- affecting the 9p21 region to which p16INK4a and p15INK4b genes have been mapped. No alterations in chromosome 9 were detected in the other four typical MCLs in which cytogenetic studies were performed.
Frequency of p53, p21Waf1, and p16INK4a Gene Alterations and Expression in MCLs
| Diagnosis . | p53 Mutations . | p21Waf1 . | p16INK4a . | ||||
|---|---|---|---|---|---|---|---|
| MCL Variant . | . | Deletions . | Mutational Analysis . | Loss of Expression . | Deletions . | Mutational Analysis . | Loss of Expression . |
| Aggressive | 2/6 | 0/5 | 0/5 | 1/6 | 3/6* | 0/3† | 2/5 |
| Typical | 0/18 | 0/18 | 1/18‡ | 0/18 | 0/18 | 1/18‡ | 0/17 |
| Total | 2/24 | 0/23 | 1/23 | 1/24 | 3/24 | 1/21 | 2/22 |
| Diagnosis . | p53 Mutations . | p21Waf1 . | p16INK4a . | ||||
|---|---|---|---|---|---|---|---|
| MCL Variant . | . | Deletions . | Mutational Analysis . | Loss of Expression . | Deletions . | Mutational Analysis . | Loss of Expression . |
| Aggressive | 2/6 | 0/5 | 0/5 | 1/6 | 3/6* | 0/3† | 2/5 |
| Typical | 0/18 | 0/18 | 1/18‡ | 0/18 | 0/18 | 1/18‡ | 0/17 |
| Total | 2/24 | 0/23 | 1/23 | 1/24 | 3/24 | 1/21 | 2/22 |
Two cases showed homozygous deletions by Southern blot analysis. One additional case had a hemizygous deletion of the 9p −del (9) (p12)−. No DNA was available in this case.
The SSCP analysis was not performed in the cases with homozygous p16INK4a gene deletion.
The two cases with anomalous SSCP had a known polymorphism in codons 31 of p21Waf1 gene and codon 148 of p16INK4a gene, respectively.
p21Waf1 and p16INK4a Gene Alterations and Expression in Aggressive MCLs
| Case . | Histology . | p53 . | p21Waf1 . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | Gene . | Expresssion . | p16INK4a . | . | |||||||
| . | . | . | SB . | SSCP/Sequence . | mRNA . | Protein (WB and IHQ) . | Gene . | Expression . | . | . | . | ||
| . | . | . | . | . | . | . | SB . | SSCP/Sequence . | mRNA . | Protein . | . | . | . |
| 3858 | Blastic | WT | +/+ | WT | + | + | −/− | NA | − | − | |||
| 15566 | Blastic | WT | +/+ | WT | ND | + | −/− | NA | ND | − | |||
| 3747 | Large cell | WT | ND | ND | + | +* | +/−† | ND | ND | ND | |||
| 441 | Blastic | WT | +/+ | WT | − | − | +/+ | WT | + | ++ | |||
| 1104 | Large cell | M Codon 273 | +/+ | WT | + | + | +/+ | WT | ++ | ++ | |||
| 4958 | Blastic | M Codon 278 | +/+ | WT | + | + | +/+ | WT | + | ND | |||
| Case . | Histology . | p53 . | p21Waf1 . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | Gene . | Expresssion . | p16INK4a . | . | |||||||
| . | . | . | SB . | SSCP/Sequence . | mRNA . | Protein (WB and IHQ) . | Gene . | Expression . | . | . | . | ||
| . | . | . | . | . | . | . | SB . | SSCP/Sequence . | mRNA . | Protein . | . | . | . |
| 3858 | Blastic | WT | +/+ | WT | + | + | −/− | NA | − | − | |||
| 15566 | Blastic | WT | +/+ | WT | ND | + | −/− | NA | ND | − | |||
| 3747 | Large cell | WT | ND | ND | + | +* | +/−† | ND | ND | ND | |||
| 441 | Blastic | WT | +/+ | WT | − | − | +/+ | WT | + | ++ | |||
| 1104 | Large cell | M Codon 273 | +/+ | WT | + | + | +/+ | WT | ++ | ++ | |||
| 4958 | Blastic | M Codon 278 | +/+ | WT | + | + | +/+ | WT | + | ND | |||
Abbreviations: SB, Southern blotting; WB, Western blot; IHQ, immunohistochemistry; SB: +/+ no deletion; +/− hemizygous deletion; −/− homozygous deletion; M, mutated; WT, wild-type; ND, not done; NA, not applicable.
In this case protein p21Waf1 protein expression was only detected by immunohistochemistry.
In this case a hemizygous deletion of 9p arm −del (9) (p12)− was detected by cytogenetic analysis. No DNA was available for Southern blotting.
Southern blot analysis of four typical and four aggressive mantle cell lymphomas. HindIII-digested DNA was electrophoresed on a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with the exon 2 of p16INK4a, p21Waf1, and bcl-1 MTC probes. The aggressive MCL 3858 and 15566 show homozygous deletions of p16INK4a and p15INK4b genes. No alterations in the p21Waf1 signal was seen in any of the cases. Case 4958 had a bcl-1 rearrangement detected with HindIII. Case 441 also had a bcl-1 rearrangement detected with EcoRI and BamHI but not with HindIII. No rearrangements were detected in the remaining cases.
Southern blot analysis of four typical and four aggressive mantle cell lymphomas. HindIII-digested DNA was electrophoresed on a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with the exon 2 of p16INK4a, p21Waf1, and bcl-1 MTC probes. The aggressive MCL 3858 and 15566 show homozygous deletions of p16INK4a and p15INK4b genes. No alterations in the p21Waf1 signal was seen in any of the cases. Case 4958 had a bcl-1 rearrangement detected with HindIII. Case 441 also had a bcl-1 rearrangement detected with EcoRI and BamHI but not with HindIII. No rearrangements were detected in the remaining cases.
To determine whether mutations of p16INK4a gene were present in these lymphomas, we analyzed exon 1 and 2 of the p16INK4a gene by PCR-SSCP. No abnormalities were detected in the 3 aggressive MCLs in which the gene was not deleted and in 17 typical MCLs. In one typical MCL (case 2166), an abnormally migrating band was identified in exon 2. Sequencing of this exon showed that the altered mobility was the result of the known polymorphism at codon 148 with the change GCG (alanine) → ACG (threonine) (Fig 2). To confirm the SSCP analysis, exons 1 and 2 of the p16INK4a gene were completely sequenced in the 3 aggressive MCLs with no p16INK4a, and in 4 additional typical MCLs. No mutations were detected in any of these cases.
SSCP analysis of p16INK4a exon 2. The abnormal mobility observed in the typical mantle cell lymphoma 2166 (A) is the result of the known polymorphism in codon 148 with the change GCG (alanine) → ACG (threonine) (B).
SSCP analysis of p16INK4a exon 2. The abnormal mobility observed in the typical mantle cell lymphoma 2166 (A) is the result of the known polymorphism in codon 148 with the change GCG (alanine) → ACG (threonine) (B).
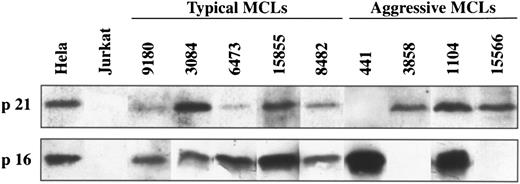
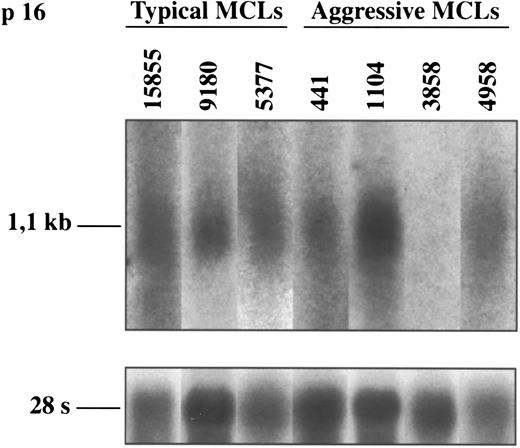
p16INK4a gene expression.To know the possible alterations of p16INK4a gene expression in these lymphomas, 5 aggressive MCLs and 17 typical variants were examined at mRNA and/or protein levels by Northern and Western blot analysis, respectively. Two of the 5 (40%) aggressive MCLs (cases 3858 and 15566) showed a complete loss of p16INK4a expression (Figs 3 and 4). In these two cases no protein was detected by Western blot (Fig 3). In addition, the lack of p16INK4a expression was confirmed by Northern blot in one of the cases (case 3858) in which RNA was available (Fig 4). p16INK4a expression at either mRNA, protein, or both protein and mRNA was detected in the other 3 aggressive MCLs and in the 17 typical variants (Figs 3 and 4). Interestingly, two other aggressive MCLs analyzed by Western blot showed a stronger overexpression of p16INK4a protein in comparison with the levels observed in the typical variants of MCLs.
Western blot analysis of p21Waf1 and p16INK4a in typical and aggressive mantle cell lymphomas. The aggressive MCL 441 shows a loss of p21Waf1 protein expression whereas MCL 3858 and 15566 have a loss of p16INK4a protein expression. These genes are constantly expressed in the typical variants of MCLs.
Western blot analysis of p21Waf1 and p16INK4a in typical and aggressive mantle cell lymphomas. The aggressive MCL 441 shows a loss of p21Waf1 protein expression whereas MCL 3858 and 15566 have a loss of p16INK4a protein expression. These genes are constantly expressed in the typical variants of MCLs.
Northern blot analysis of p16INK4a in typical and aggressive mantle cell lymphomas. The aggressive MCL 3858 shows a loss of p16INK4a mRNA expression. This finding is concordant with the lack of protein expression and the homozygous deletion of the gene in this case (shown in Figs 1 and 3, respectively). The blots were stripped of the p16INK4a probe and rehybridized with the 28S probe as a loading control.
Northern blot analysis of p16INK4a in typical and aggressive mantle cell lymphomas. The aggressive MCL 3858 shows a loss of p16INK4a mRNA expression. This finding is concordant with the lack of protein expression and the homozygous deletion of the gene in this case (shown in Figs 1 and 3, respectively). The blots were stripped of the p16INK4a probe and rehybridized with the 28S probe as a loading control.
The two aggressive cases with lack of p16INK4a expression were the two cases with gene deletions detected by Southern blot. Unfortunately, p16INK4a expression could not be analyzed in the aggressive MCL with the 9p12 hemizygous deletion detected by cytogenetic analysis because not enough RNA was available (p16INK4a mRNA message was only clearly shown in Northern blots with 20 μg of total RNA) and no additional frozen material was available from this patient.
p21Waf1 gene analysis.p21Waf1 gene structure was analyzed by Southern blot analysis and SSCP in the 18 typical and 5 aggressive MCLs (Tables 1 and 2). No abnormalities in p21Waf1 gene were detected by Southern blot analysis in any of the typical or aggressive variants of MCLs (Fig 1).
Exon 2 of p21Waf1 gene was also examined by PCR-SSCP to screen for possible mutations. No altered mobility was observed in any of the 5 aggressive MCLs and in 17 typical MCLs. In one typical case (case 9180) an abnormal band was detected. Direct sequencing of the fragment with aberrant mobility showed the known polymorphism in codon 31 with the change AGC (serine) → AGA (arginine). To confirm the SSCP analysis, the full coding sequence (exon 2 and 3) of the p21Waf1 gene was completely sequenced in 4 aggressive MCLs and 4 additional typical MCLs. No mutations were detected in any of these cases.
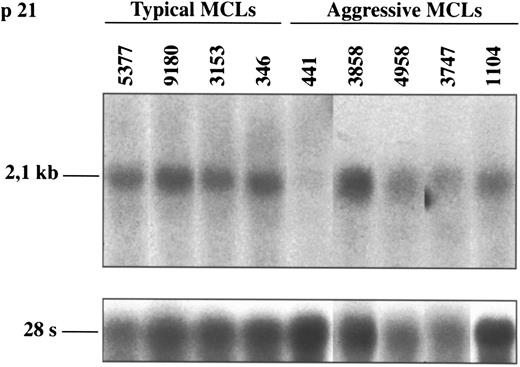
p21Waf1 gene expression.p21Waf1 expression was analyzed by Northern and/or Western blot in the 6 aggressive MCLs and in 18 typical variants. One aggressive MCL (case 441) showed a loss of p21Waf1 expression at both mRNA and protein level (Figs 3 and 5). p21Waf1 protein and/or mRNA expression was detected in the remaining 5 aggressive MCLs and in all typical cases.
Northern blot analysis of p21Waf1 in typical and aggressive mantle cell lymphomas. The aggressive MCL 441 shows a loss of p21Waf1 mRNA expression. This finding is concordant with the lack of protein expression in this case (shown in Fig 3). The blots were stripped of the p16INK4a probe and rehybridized with the 28S probe as a loading control.
Northern blot analysis of p21Waf1 in typical and aggressive mantle cell lymphomas. The aggressive MCL 441 shows a loss of p21Waf1 mRNA expression. This finding is concordant with the lack of protein expression in this case (shown in Fig 3). The blots were stripped of the p16INK4a probe and rehybridized with the 28S probe as a loading control.
p21Waf1 could also be shown by immunohistochemical staining of formalin-fixed and paraffin-embedded material using an antigen retrieval technique. p21Waf1 expression was detected in all typical MCLs with a nuclear pattern. No cytoplasmic expression was observed in any case. The number of stained cells was usually scarce, varying from 1.5% to 5.2% (2.8% ± 1.4%, mean ± SD) of the cells. In the aggressive variants, immunostaining was similarly detected in the nucleus of the cells. The number of positive cells was higher than in typical variants (5.5 ± 1.6; mean ± SD). However, no positive cells were observed in the aggressive MCL in which no expression had been detected by Western or Northern blot analysis.
Correlation between p53 mutations and p21Waf1/p16INK4a gene alterations.The comparison between p53 mutations, p16INK4a, and p21Waf1 gene alterations in the 6 aggressive MCLs is summarized in Table 2. p53 gene mutations were detected in two of the 6 aggressive MCLs. The mutations were found at codon 278 (case 4958) and codon 273 (case 1104). Interestingly, p21Waf1 expression could be shown in both cases at mRNA level. In one of these cases (case 1104), p21Waf1 expression was also confirmed at protein level by Western blot analysis. The only aggressive MCLs in which no p21Waf1 expression could be detected showed a wild-type p53. No p53 expression was detected in this case by immunohistochemistry. To confirm the SSCP-negative analysis in this case, exons 4 to 10 were completely sequenced and no mutations were detected. Wild-type p53 was also found in the two aggressive MCLs with loss of p16INK4a expression and in the aggressive MCL with hemizygous deletion of 9p12. Therefore, all aggressive MCLs in this series were molecularly characterized by alterations in one of these 3 genes: p53 mutations in 2 cases, p16INK4a gene deletions and loss of expression in 2 cases, and p21Waf1 loss of expression in 1 case. One additional aggressive MCL had a hemizygous deletion of 9p12. No alterations in any of these genes were observed in typical variants of MCLs.
DISCUSSION
In this study we have examined the gene structure and expression of the p16INK4a and p21Waf1 CDKIs genes in 18 typical and 6 aggressive variants of MCLs. All these cases had been previously characterized for the expression of cyclin D1, retinoblastoma status, and the presence of p53 mutations.8,28 34 Deletions and/or loss of expression of p16INK4a and p21Waf1 genes were found only in 4 of the 24 (17%) MCLs. However, the four cases were in the histologically aggressive subgroups, further classified as blastic or large cell variants of MCL. mRNA and/or protein expression were detected in all typical MCLs with low proliferative activity and longer survival. Concordantly, no gene deletions were observed in any of these typical MCLs. These findings indicate that alterations of the CDKI p16INK4a and p21Waf1 genes in MCLs are a relatively infrequent phenomenon (17% of all the MCLs examined). However, the presence of deletions and loss of expression in 4 of the 6 histologically aggressive cases (67%) with higher proliferative activity and shortened survival indicates that inactivation of these CDKIs genes may be involved in the pathogenesis of a subset of aggressive MCLs. All of our aggressive MCLs were diagnosed at presentation, suggesting that these molecular alterations may occur in early phases of the evolution of these lymphomas. Aggressive variants may also evolve as a progression of typical MCLs. It will be interesting to investigate the possible implication of CDKIs genes in the aggressive transformation of more indolent typical MCL variants.
Homozygous deletions of p16INK4a and p15INK4b CDKIs genes are a common genetic event in acute lymphoblastic leukemias (ALL), particularly of T-cell phenotype. Several studies have reported homozygous deletions of the p16INK4a gene in 18% to 83% of T-ALL and in 8% to 21% of B-ALL.44,47-50 The status of these genes in non-Hodgkin lymphomas (NHLs) has been less well examined, but gene deletions seem to be an infrequent phenomenon in these neoplasms, ranging from 0% to 7% of the cases. However, most of these deletions have been described in diffuse large cell lymphomas de novo or in the transformed phase of a low-grade NHL.50,51 Our findings in MCLs are concordant with these studies in other NHL. Deletions of p16INK4a/p15INK4b genes or the 9p21 chromosomal region where these genes are mapped were found in 3 of the 6 aggressive variants of MCL (50%) but in none of the typical MCL. Koduru et al51 have recently examined the presence of p16INK4a/p15INK4b gene deletions in a large series of NHL including 13 cases with the t(11; 14)(q13; q32). A deleted p16INK4a/p15INK4b gene was found in 4 of the 13 lymphomas with the t(11; 14) (31%), but only in 9 of 141 NHL (6%) with other karyotypic abnormalities or normal karyotypes. In this study, the NHLs were classified according to the Working Formulation and, consequently, the diagnosis of mantle cell lymphoma was not considered in any of the cases. Interestingly, the four cases with the t(11; 14) and deletions of p16INK4a/p15INK4b genes were classified as diffuse large cell lymphomas. The other 9 cases with the t(11; 14) and no deletions of the p16INK4a/p15INK4b genes were classified as other low- or intermediate-grade categories of the Working Formulation. Because the t(11; 14) translocation is now recognized as a genetic alteration highly characteristic of MCL and it is only very infrequently found in other NHLs,52 it is possible that these cases with the t(11; 14) and a deleted p16INK4a/p15INK4b gene may represent aggressive variants of MCLs. These observations and the findings in our study would suggest that deletions of p16INK4a/p15INK4b genes may be an important genetic event in the pathogenesis of aggressive variants of MCLs.
The p16INK4a gene expression in lymphoproliferative disorders has been less well analyzed than the structure of the gene. In our study, p16INK4a expression was observed at mRNA and/or protein levels in all typical MCLs. However, no expression of the gene was detected in two (40%) aggressive variants with a blastic morphology. The Southern blot analysis of these two cases showed a homozygous deletion of p16INK4a gene. The loss of p16INK4a expression in the two blastic MCLs confirms the implication of this gene in the development of these aggressive variants of MCLs.
The levels of p16INK4a expression in two other aggressive MCLs were much higher than the levels detected in the typical MCLs. High levels of p16INK4a expression have been detected in cell lines with lack of pRb function. Several studies have provided evidences for a regulatory loop between p16INK4a expression, retinoblastoma protein, and cyclin D/cdks complexes.53,54 In this model, transcription of p16INK4a would be repressed by unphosphorylated pRb whereas phosphorylated pRb would activate p16INK4a transcription. Concordantly with this idea, we have shown in a previous study that the two aggressive MCLs with p16INK4a overexpression showed high levels of phosphorylated pRb whereas the typical MCLs constantly expressed low levels of pRb with negligible phosphorylated forms.34 The inefficient inhibitory activity of p16INK4a in these cases may be caused by alterations in other inhibitory molecules. In fact, the two aggressive cases with p16INK4a overexpression had a loss of p21Waf1 expression and a p53 mutation, respectively.
The p21Waf1 gene is regulated by p53 and can suppress the growth of human normal and tumor cells by inhibiting the activity of cyclins/CDKs complexes.18,19 Overexpression of p21Waf1 has been shown to suppress proliferation and tumorigenicity in several human cell lines including Burkitt's lymphoma cells.21,22 The possible role of p21Waf1 gene alterations in the pathogenesis of human lymphoproliferative disorders has not been well examined. In this study, p21Waf1 protein and mRNA expression were detected in all typical MCLs. However, one aggressive MCL with a blastic morphology showed a loss of both mRNA and protein expression, suggesting that abrogation of p21Waf1 expression may be implicated in the higher proliferative activity of this tumor. The other 5 aggressive MCLs showed similar levels of expression as the typical variants. p21Waf1 expression in these lymphomas seems to be independent of the status of the p53 gene. No mutations were found in exons 4 to 10 in the blastic MCL with no expression of p21Waf1. On the other hand, two aggressive MCLs with p53 mutations showed similar levels of p21Waf1 expression at mRNA and protein levels as the tumors with wild-type p53. Several studies have now shown p53-independent induction of p21Waf1 expression, particularly in the terminal differentiation process of several cell lineages20,55,56 and in response to different growth factors.57 The presence of p21Waf1 expression in two aggressive MCLs with p53 mutations suggests that other p53-responsive genes may be involved in the pathogenesis of these tumors. No p21Waf1 gene deletions or mutations were detected in our blastic MCL with no p21Waf1 expression, indicating that other mechanisms must be implicated in the inactivation of the expression of this gene.
The comparative analysis of alterations in our series of MCLs shows that these molecular abnormalities occur independently of each other. The independent occurrence of p53, p16INK4a, and p21Waf1 gene alterations supports the idea that inactivation of any one of these genes is sufficient to induce a higher proliferative activity of the tumors. p16INK4a, cyclin D1, and retinoblastoma protein seem to participate in the same cell-cycle regulatory pathway.30-33 Inactivation of p16INK4a expression and overexpression of cyclin D1 cooperate synergistically to deregulate cell proliferation in cell lines with normal pRb function.33 Our observations would suggest that p53 mutations and p21Waf1 downregulation may also cooperate with cyclin D1 overexpression in cell-cycle deregulation in tumors with a normal pRb. The constant overexpression of cyclin D1 in MCLs has been considered to play an important role in the pathogenesis of these lymphomas. The findings of this study would indicate that the additional alteration in one of the negative cell-cycle regulatory genes p53, p16INK4a, or p21Waf1 may lead to development of more aggressive variants of these tumors with higher proliferative activity and shorter survival of the patients.
ACKNOWLEDGMENT
The authors thank Dr M. Serrano for the gift of the p16INK4a probe and his comments on the manuscript, and Iracema Nayach and Nerea Peiro for excellent technical assistance.
Supported in part by Grants No. SAF 1195/93, SAF 96/61 from CICYT, and F1596/236 Ministerio de Educación y Ciencia (E.C.). M.P. and L.H. were fellows supported by a grant from the European Union; M.C. and P.J. were fellows from the Spanish Ministerio de Educación y Ciencia.
The first and second authors contributed equally to this study.
Address reprint requests to Elı́as Campo, MD, Laboratory of Anatomic Pathology, Hospital Clinic Provincial, Villarroel 170, 08036-Barcelona, Spain.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal