Abstract
CRKL has previously been shown to be a major tyrosine phosphorylated protein in neutrophils of patients with BCR-ABL+ chronic myelogenous leukemia and in cell lines expressing BCR-ABL. CRKL and BCR-ABL form a complex as demonstrated by coimmunoprecipitation and are capable of a direct interaction in a yeast two-hybrid assay. We have mapped the site of interaction of CRKL and BCR-ABL to the amino terminal SH3 domain of CRKL with a proline rich region in the C-terminus of ABL. The proline-rich region was mutated and the effect of this deletion on BCR-ABL transforming function was assayed. Our data show that this deletion does not impair the ability of BCR-ABL to render myeloid cells factor independent for growth. In cells expressing the proline deletion mutation of BCR-ABL, CRKL is still tyrosine phosphorylated and forms a complex with BCR-ABL as demonstrated by coimmunoprecipitation. Our data suggest that the interaction between CRKL and the proline deletion mutant of BCR-ABL is an indirect interaction as CRKL does not interact directly with the proline deletion mutant of BCR-ABL in a gel overlay assay or in a yeast two-hybrid assay. Thus, a direct interaction of CRKL and BCR-ABL is not required for CRKL to become tyrosine phosphorylated by BCR-ABL and suggests that CRKL function may still be required for BCR-ABL function through an indirect interaction.
CHRONIC MYELOGENOUS leukemia (CML) is a hematopoietic stem cell malignancy that constitutes approximately 20% of cases of adult leukemia. In more than 95% of patients with CML, a reciprocal translocation between chromosomes 9 and 22, known as the Philadelphia chromosome translocation is present.1,2 This balanced translocation juxtaposes the breakpoint cluster region (BCR) from chromosome 22 with the c-ABL proto-oncogene on the long arm of chromosome 9.3-5 The BCR-ABL fusion creates a gene whose protein product has tyrosine kinase activity increased several fold over the normal c-ABL gene product.6-8
The BCR-ABL fusion protein has been shown to transform hematopoietic progenitor cells in bone marrow (BM) culture,9-11 to transform interleukin-3 (IL-3)–dependent myeloid cell lines to growth factor independence,12,13 and to cause a syndrome resembling chronic myelogenous leukemia in syngeneic mice.14,15 Although ABL kinase activity has been shown to be required for transformation of myeloid cell lines by P210 BCR-ABL,8,16 the mechanism by which transformation takes place is not yet known. Although numerous substrates of the BCR-ABL tyrosine kinase have been identified, the requirement of most of these substrates for the transforming function of BCR-ABL is similarly unknown. To assist in determining which of the substrates of BCR-ABL may be physiologically relevant for BCR-ABL function, we compared CML patient samples to normal controls for differences in tyrosine phosphorylated proteins. Through this analysis, we and others have shown that the SH2-SH3 adaptor protein, CRKL, is heavily tyrosine phosphorylated in neutrophils of patients with Philadelphia chromosome-positive CML.17-19
CRKL is a 39-kD protein that is related to the CRK oncogene of the avian sarcoma virus, CT10.20 Two human homologs of CRK, in addition to CRKL, have been identified, termed CRK I and CRK II, that differ by the presence of a second SH3 domain in CRK II as a result of alternative splicing.21 CRK II is most similar to CRKL in that both contain two SH3 domains. CRKL is not only tyrosine phosphorylated in CML patient samples, but it is also tyrosine phosphorylated in cell lines that express BCR-ABL and is inducibly phosphorylated in a BCR-ABL temperature-sensitive mutant.17,22,23 CRKL and BCR-ABL form a complex as shown by coimmunoprecipitation and we have previously demonstrated direct association of CRKL with BCR-ABL sequences in the yeast two-hybrid system.17 These studies suggest a significant role for this adaptor protein in BCR-ABL transformation.
Therefore, in the current study we have mapped the direct binding site between CRKL and ABL. BCR-ABL mutants with a deletion of the CRKL binding site were constructed and analyzed for transforming abilities. Transforming function of BCR-ABL was preserved in this deletion mutant, but surprisingly, tyrosine phosphorylation of CRKL and persistent coimmunoprecipitation of CRKL with the BCR-ABL deletion mutant was observed. Although our data demonstrate that we have abolished direct interaction between BCR-ABL and CRKL, we show that CRKL is also capable of associating with BCR-ABL indirectly, through another protein. Thus, we cannot exclude a significant role for CRKL in transformation by BCR-ABL via indirect interactions.
MATERIALS AND METHODS
Generation of glutathione-S-transferase (GST)-Fusion Constructs and Mutants
A schematic of the GST-CRKL fusion proteins is depicted (see Fig 2). GST-CRKL SH2 was made by excising an Xba I to Ecl 136 II fragment from pGEX-CRKL17 and replacing it with an Xba I to Rsa I fragment from the same plasmid. CRKL SH3 was obtained by polymerase chain reaction (PCR) using the primers: 5′-ACAGCAGAAGATAACCTG-3′ and 5′-GTATCTCGCCATGGCCTCAAG-3′. The PCR product was digested with Nco I and ligated into pGEX-KG that had been digested with EcoRI, filled in with Klenow (New England Biolabs, Beverly, MA) and Nco I digested. pGEX-CRKL SH3n was constructed by digesting pGEX-CRKL-SH3 with HindIII and religating. CRKL SH3c was constructed by PCR using the following primers: 5′-GTCTTTGCGAAAGCAATC-3′ and 5′-GTATCTCGCCATGGCCTCAAG-3′. The PCR product was digested with Nco I and ligated into pGEX-KG prepared as for the CRKL SH2 construct. The intra SH3 domain was prepared by PCR using the primers: 5′-ACAGCAGAAGATAACCTG-3′ and 5′-CCCAAGCTTTTCTGGATTGCTTTCGC-3′. The PCR product was digested with HindIII and ligated into HindIII-digested pGEX-KG. The FLVRES mutation, a substitution mutation of Arg to Lys at amino acid 39, was created by single strand mutagenesis using the Amersham Sculptor mutagenesis system (Amersham, Arlington Heights, IL) according to the manufacturer's instructions. Single-strand DNA was made from CRKL cloned as an Xba I to Xho I fragment into pBD3.24 The oligonucleotide used for mutagenesis was: R-K39CRKL, 5′-CCTCGTCAAGGATTCTTCCACC-3′. CRK II was obtained by reverse transcriptase (RT)-PCR from HeLa cells using the following primers: 5′-GAGGCAGCCATGGCGGGCAAC-3′ and 5′-AGATTGTTCCCATCTGTCAGC-3′ and was cloned into pGEX-KG. All of the above constructs were sequenced to verify that no mutations had been introduced by the PCR reactions and that no second-site mutations had been introduced during mutagenesis.
Schematic of CRKL constructs. Full-length and various portions of CRKL were subcloned into pGEX-KG as described in Materials and Methods and are depicted in this figure.
Schematic of CRKL constructs. Full-length and various portions of CRKL were subcloned into pGEX-KG as described in Materials and Methods and are depicted in this figure.
ABL Deletion Mutants
Deletion of two proline rich sequences from the C-terminus of ABL (see Fig 1) was accomplished by PCR mutagenesis on a fragment of ABL corresponding to an AatII to Bcl I fragment using the following primers: 5′ ACCTCCAGGAGAGCTGCAGAG-3′ and 5′-GGCCTGCAGCAAGGTAGTCAC-3′ for deletion of the site referred to as P1 in Fig 1 and 5′-GAGCGAGGTCCCCCGGAGG-3′ and 5′-AGACACGGCAGGCTCATGGTC-3′ for the site referred to as P2 in Fig 1. The double mutant containing a deletion of both sites was obtained by deleting the second site from a plasmid containing a deletion of the first site. The AatII to Bcl I fragment was sequenced to ensure that no additional mutation had been obtained and was repackaged into full length BCR-ABL in pUC9 that had been modified to remove the existing AatII site.16 The resulting BCR-ABL mutant was cloned into the retroviral vector pGD15 (gift of G. Daley, MIT, Cambridge, MA).
Schematic of ABL construct used in the yeast two-hybrid screen. Numbers correspond to amino acids numbered according to c-ABL type Ia. Arg 271 corresponds to the substitution at this amino acid that yields a kinase defective ABL. P1 and P2 are two proline-rich regions in the C-terminus of ABL.
Schematic of ABL construct used in the yeast two-hybrid screen. Numbers correspond to amino acids numbered according to c-ABL type Ia. Arg 271 corresponds to the substitution at this amino acid that yields a kinase defective ABL. P1 and P2 are two proline-rich regions in the C-terminus of ABL.
Antisera
Rabbit polyclonal antisera against CRKL were generated against a peptide corresponding to amino acids 204 to 225 of CRKL. This antisera was used exclusively for immunoblotting experiments. For other experiments, anti CRKL antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies recognizing p120 CBL, p145 C3G, and ABL proteins (24-11 and K-12) were purchased from Santa Cruz Biotechnology. The K-12 ABL antibody was used for some of the immunoprecipitation experiments and was found to more efficiently precipitate BCR-ABL than c-ABL. Antibodies recognizing the 85-kD subunit of PI-3 kinase, GRB-2, SHC, p120 rasGAP, and SOS were obtained from UBI (Lake Placid, NY). Anti p130 CAS was obtained from Transduction Laboratories (Lexington, KY). The paxillin monoclonal antibody (MoAb) was a generous gift of Drs J. Griffin and R. Salgia (Dana-Farber Cancer Institute, Boston, MA). The antiphosphotyrosine MoAb, 4G10, was generated using KLH-phosphotyramine as the immunogen and was used as described.25 26
Cell Lines
The 32Dcl3 cell line27 was obtained from Joel Greenberger (University of Massachusetts Medical Center, Worcester). Cells were cultured in RPMI 1640 medium (GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum (UBI), and 15% WEHI-3B conditioned media as a source of interleukin-3 (IL-3). Control retrovirus without insert, pGD, a full-length BCR-ABL, a tyrosine kinase–defective mutant of BCR-ABL,16 and the BCR-ABL deletion mutants were introduced into the 32Dcl3 cells by electroporation as described.23 Two days after transfection cells were selected in the presence of 1 mg/mL of G418 (GIBCO) in the presence of 15% WEHI-3B conditioned media. Individual clones were selected after growth in soft agar and analyzed for IL-3–independent growth.
Immunoprecipitation and Immunoblotting
32D, 32Dp210, and 32Dp210ΔP1,P2 cells were lysed in NP40 lysis buffer (20 mmol/L Tris, pH 8.0, 1 mmol/L EDTA, 150 mmol/L NaCl, 1% NP40, 10% glycerol) containing 10 μg/mL aprotinin, 1 mmol/L Na3VO4 , and 1 mmol/L phenylmethylsulfonyl fluoride. Lysates were normalized in preliminary experiments by immunoblotting for c-ABL and these amounts were used for subsequent experiments. Normalized lysates were either immunoprecipitated with 25 μL of CRKL or ABL antisera followed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or were analyzed as whole-cell lysates by SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride (PVDF) (Immobilon-P; Millipore, Bedford, MA) membranes in a buffer containing 25 mmol/L Tris, 192.5 mmol/L glycine, 20% MeOH for 4 hours at 0.45 A. Residual binding sites were blocked by incubation in TBS (10 mmol/L Tris, pH 8.0, 150 mmol/L NaCl) containing 3% bovine serum albumin (BSA) for 60 minutes at 25°C. The blots were incubated for 4 to 16 hours at room temperature with primary antibody. Antibody reactions were detected using enhanced chemiluminescence (Pierce, Rockford, IL).
GST Binding Assays
GST-fusion constructs were expressed in isopropylthio-β-D-galactosidase (IPTG)-induced Escherichia coli (DH-5α). The GST-fusion proteins were isolated from sonicated bacterial lysates using glutathione Sepharose beads (Pharmacia, Piscataway, NJ). Coomassie-stained gels of the GST-fusion proteins were used to normalize for the expression of the various GST-fusion proteins. Between 2.5 and 5 μg of GST-fusion proteins were incubated with 50 μL of glutathione Sepharose beads in bacterial lysis buffer (150 mmol/L NaCl, 16 mmol/L Na2HPO4 , 4 mmol/L NaH2PO4 , [pH 7.3] containing 10 μg/mL aprotinin, 1 mmol/L Na3VO4 , 1 mmol/L phenylmethylsulfonylfluoride, and 0.1% β-mercaptoethanol). The beads were washed four times in ice-cold phosphate-buffered saline (PBS) and incubated for 4 hours with normalized cell lysates. The beads were washed three times with PBS and boiled in SDS-sample buffer. Proteins were separated by SDS-PAGE and transferred onto PVDF membranes for immunoblot analysis.
Gel Overlay Assay
Purified GST, GST-CRKL, GST-CRKL SH3n, or GST-CRKL SH2 fusion proteins were used for this assay. PVDF membranes were blocked overnight at 4°C in PBS with 0.05% Tween 20 and 5% nonfat dry milk. The blots were washed twice in PBS with 0.05% Tween 20 (PBS-T) and incubated for 2 hours at room temperature with either GST only or GST-CRKL fusion proteins (at 2 μg/mL) in binding buffer (25 mmol/L Na-phosphate [pH 7.20], 150 mmol/L NaCl, 0.1% Tween 20, 2.5 mmol/L EDTA, 20 mmol/L NaF, 1% nonfat milk, 1 mmol/L dithiothreitol, 10 μg/μL leupeptin, and 10 μg/mL aprotinin). Bound GST protein was detected by incubation with anti-GST antibody (Santa Cruz Biotechnology) diluted 1:500 in binding buffer excluding milk, dithiothreitol (DTT), and protease inhibitors. Antibody reactions were developed using enhanced chemiluminescence. The blots were washed for 1 hour in PBS-T between each of the steps.
Cell Proliferation Assays
MTT assay.Cells, 2 × 104, were plated in quadruplicate into 96-well plates in RPMI 1640 medium with 10% fetal bovine serum (FBS) with or without 15% WEHI-3B conditioned media as a source of IL-3. Wells were assayed for MTT uptake at 24-hour intervals as described.28 For each assay, controls were performed using serial dilutions of 32D and 32Dp210 cells to ensure that assay was linear with respect to cell number over the time period examined. Results are presented as the mean absorbance at 570 nm (optical density [OD]) ± standard deviation.
Cell count assays.Cells, 2 × 105, were plated in media with or without 15% WEHI-3B conditioned media. Viable cells, as assayed by the exclusion of trypan blue, were counted at daily intervals.
Yeast Two-Hybrid Assay
Analysis of interactions using the yeast two-hybrid system were performed as described.29 The ABL construct included a Kpn I to Bcl I fragment of c-ABL corresponding to the SH2, tyrosine kinase and a small portion of the C-terminal domain fused in frame to the DNA binding domain of LexA (Fig 1). The proline deletion mutants described above and a kinase inactive version of ABL16 were cloned into this ABL construct. A full-length p185BCR-ABL (gift of O. Witte, UCLA, Los Angeles, CA) was also expressed as a LexA fusion protein. Full-length CRKL and the amino terminal SH3 domains of CRKL and CRK II were expressed in frame fused to the acidic activation domain VP16. Expression of each of these constructs was confirmed by immunoblotting of yeast lysates with the appropriate antisera. Interaction was identified by selection on plates lacking histidine and analyzed for β-galactosidase production as described.29 Controls included plasmid, Lex A, VP-16, and LexA-lamin controls.
RESULTS
Mapping the Site of Interaction Between CRKL and ABL
We previously had identified CRKL and CRKII as proteins that interact with ABL in a yeast two-hybrid screen.17 Several factors suggested that an SH3 domain of CRKL was interacting with a proline-rich sequence of ABL. All of the CRKL and CRKII clones that were sequenced clustered around the amino terminal SH3 domain of these proteins. Both CRKL and CRKII bound to a kinase inactive mutant of ABL, suggesting that this was not a phosphotyrosine-dependent interaction with the SH2 domain of CRKL. Ren et al30 had previously mapped the CRKII binding site to a proline-rich region just 3′ of the kinase domain of ABL. This proline-rich region was included in the construct that we had used for our yeast two-hybrid screen and is shown schematically in Fig 1.
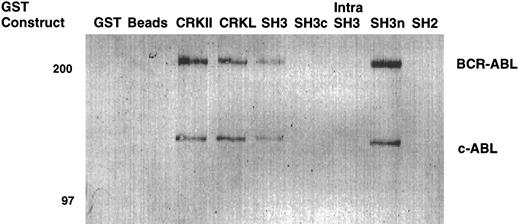
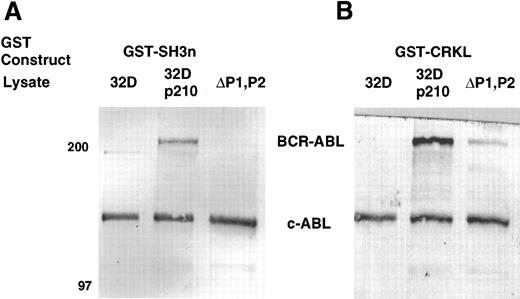
To confirm that the amino terminal SH3 domain of CRKL was interacting with ABL, various portions of CRKL were expressed as GST fusion proteins. These constructs are depicted in Fig 2. Lysates of BCR-ABL–expressing cells were allowed to bind to these GST fusion constructs. These experiments demonstrated that both c-ABL and BCR-ABL bound to full-length CRKL and that this binding was mediated predominantly by the amino terminal SH3 domain of CRKL (Fig 3). On longer exposure of the gels, there appeared to be trace amounts of binding of BCR-ABL to the SH2 domain of CRKL. There was no binding of ABL or BCR-ABL to SH3c, and in the construct containing both SH3c and SH3n (SH3) it appears that the presence of SH3c decreases the amount of ABL proteins that bind.
Binding of ABL proteins to bacterially expressed CRKL. Lysates from 1 × 107 32Dp210 cells were analyzed for binding to GST, glutathione beads (beads), or the various GST-CRKL constructs as described in Materials and Methods. Bound proteins were separated by SDS-PAGE and transferred to PVDF membranes. ABL proteins were detected using the 24-21 ABL MoAb. Molecular weight markers are indicated on the left side of the panel.
Binding of ABL proteins to bacterially expressed CRKL. Lysates from 1 × 107 32Dp210 cells were analyzed for binding to GST, glutathione beads (beads), or the various GST-CRKL constructs as described in Materials and Methods. Bound proteins were separated by SDS-PAGE and transferred to PVDF membranes. ABL proteins were detected using the 24-21 ABL MoAb. Molecular weight markers are indicated on the left side of the panel.
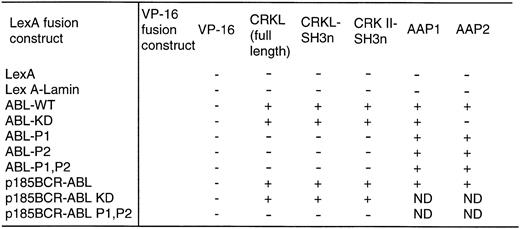
With the above information, two proline-rich sequences contained in the C-terminal region of c-ABL (referred to as P1 and P2 in Fig 1) were deleted individually or in combination. Each of these constructs was expressed as a fusion protein containing the DNA-binding domain of LexA in a yeast plasmid and analyzed for interaction with either full-length CRKL or the amino terminal SH3 domain of CRKL, expressed in frame, fused to the acidic activation domain, VP16. As shown in Fig 4, deletion of either proline-rich sequence abolished the interaction of ABL with CRKL.
Binding of CRKL to ABL in the yeast two-hybrid system. ABL and BCR-ABL constructs and a lamin control were expressed as LexA-fusion proteins and analyzed for interaction with CRKL and CRK II expressed as VP-16 acidic activation domain fusion proteins. Two additional VP-16 constructs, AAP1 and AAP2, that were identified in our yeast two-hybrid screen using the ABL kinase domain were used as controls. + indicates positivity for β-galactosidase activity and growth on his− plates. Abbreviations: ABL-WT refers to the SH2, kinase construct depicted in Fig 1; ABL-KD is a kinase defective version of ABL; P1, P2, are the deletion mutants of the proline-rich regions depicted in Fig 1.
Binding of CRKL to ABL in the yeast two-hybrid system. ABL and BCR-ABL constructs and a lamin control were expressed as LexA-fusion proteins and analyzed for interaction with CRKL and CRK II expressed as VP-16 acidic activation domain fusion proteins. Two additional VP-16 constructs, AAP1 and AAP2, that were identified in our yeast two-hybrid screen using the ABL kinase domain were used as controls. + indicates positivity for β-galactosidase activity and growth on his− plates. Abbreviations: ABL-WT refers to the SH2, kinase construct depicted in Fig 1; ABL-KD is a kinase defective version of ABL; P1, P2, are the deletion mutants of the proline-rich regions depicted in Fig 1.
Effects of the Proline Deletion on BCR-ABL Transformation
To analyze the effects of deletion of the site of interaction of CRKL with BCR-ABL, the proline deletion mutants were repackaged into full-length BCR-ABL. As the results of the deletion of each individual proline-rich region and the double mutant are identical, only the results of the double mutant, referred to as p210ΔP1,P2 will be presented. An IL-3–dependent murine cell line, 32D, was transfected with full-length p210BCR-ABL, p210ΔP1,P2, and control retrovirus and selected in the presence of neomycin and IL-3. Individual clones were selected after growth in soft agar and were analyzed for IL-3–independent growth. Figure 5 is representative of results obtained from 10 of 10 clones expressing BCR-ABL and shows that the p210ΔP1,P2-expressing cells are capable of IL-3–independent growth. Similarly, there was no defect in fibroblast transformation by this mutant (data not shown). Analysis of BCR-ABL kinase activity showed no difference between the proline deletion mutant and the full-length version of p210BCR-ABL (data not shown).
Proliferation assay of BCR-ABL ΔP1,P2 mutant in 32D cells. 32D, 32Dp210, and 32Dp210ΔP1,P2 cells growing in the presence of IL-3 were washed three times with RPMI 1640 medium and plated at 1 × 105 cells/mL in regular growth media with or without exogenous growth factor. Each day, viable cells were counted as assessed by exclusion of trypan blue. The data presented are representative of four separate experiments. Similar results were obtained in MTT assays. (▪), 32D + IL-3; (□), 32D − IL-3; (•), 32Dp210 + IL-3; (○), 32Dp210 − IL-3; (▴), 32Dp210(ΔP1,P2) + IL-3; (▵), 32Dp210(ΔP1,P2) − IL-3.
Proliferation assay of BCR-ABL ΔP1,P2 mutant in 32D cells. 32D, 32Dp210, and 32Dp210ΔP1,P2 cells growing in the presence of IL-3 were washed three times with RPMI 1640 medium and plated at 1 × 105 cells/mL in regular growth media with or without exogenous growth factor. Each day, viable cells were counted as assessed by exclusion of trypan blue. The data presented are representative of four separate experiments. Similar results were obtained in MTT assays. (▪), 32D + IL-3; (□), 32D − IL-3; (•), 32Dp210 + IL-3; (○), 32Dp210 − IL-3; (▴), 32Dp210(ΔP1,P2) + IL-3; (▵), 32Dp210(ΔP1,P2) − IL-3.
Analysis of CRKL Phosphorylation and Interaction With BCR-ABL
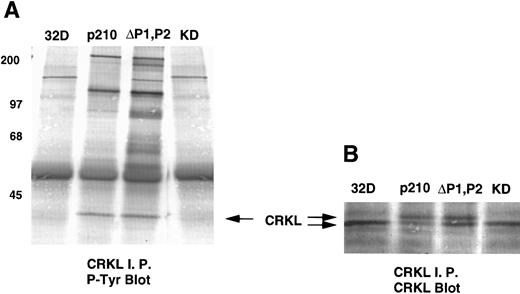
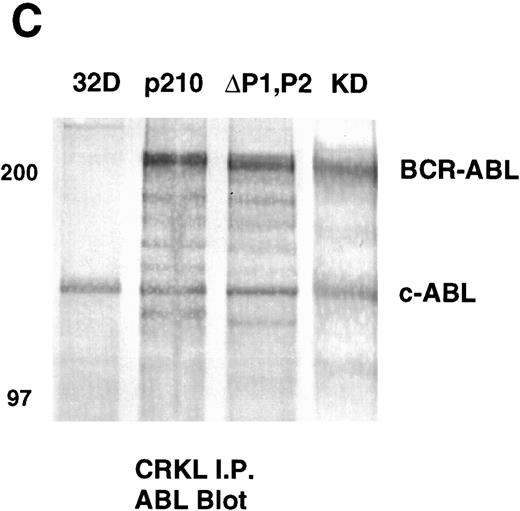
Analysis of whole-cell lysate phosphotyrosine immunoblots of cells expressing the p210ΔP1,P2 mutant showed a pattern of tyrosine phosphorylated proteins that was similar to that of cellular lysates obtained from cells expressing the full-length BCR-ABL. This included prominent tyrosine phosphorylation of a 39-kD protein that comigrated with the 39-kD protein that we had previously purified from K562 cells and had shown to be CRKL.17 That CRKL was tyrosine phosphorylated in the p210ΔP1,P2-expressing cells was confirmed by CRKL immunoprecipitates followed by antiphosphotyrosine immunoblotting (Fig 6A). Further, CRKL and BCR-ABL remained capable of interacting as demonstrated by coimmunoprecipitation. Thus, CRKL immunoprecipitates from lysates of cells expressing the p210ΔP1,P2 mutant contain BCR-ABL and ABL (Fig 6C). Similar results for CRKL and BCR-ABL association were obtained using a p185 construct containing a similar deletion of the proline-rich sequences (data not shown). A kinase-defective BCR-ABL also associates with CRKL, consistent with binding of CRKL to a region outside of the tyrosine kinase domain of ABL. However, there is no tyrosine phosphorylation of CRKL in cells expressing the kinase defective version of BCR-ABL.
Tyrosine phosphorylation of CRKL and coimmunoprecipitation of BCR-ABL and CRKL. Lysates of cells expressing the indicated constructs were (A) immunoprecipitated with CRKL antisera and immunoblotted with 4G10, a monoclonal antiphosphotyrosine antibody; (B) immunoprecipitated with CRKL antisera and immunoblotted with CRKL antisera; (C) immunoprecipitated with CRKL antisera and immunoblotted with an ABL MoAb. KD refers to a tyrosine kinase defective mutant of BCR-ABL. Molecular weight markers are indicated on the left side of the panel.
Tyrosine phosphorylation of CRKL and coimmunoprecipitation of BCR-ABL and CRKL. Lysates of cells expressing the indicated constructs were (A) immunoprecipitated with CRKL antisera and immunoblotted with 4G10, a monoclonal antiphosphotyrosine antibody; (B) immunoprecipitated with CRKL antisera and immunoblotted with CRKL antisera; (C) immunoprecipitated with CRKL antisera and immunoblotted with an ABL MoAb. KD refers to a tyrosine kinase defective mutant of BCR-ABL. Molecular weight markers are indicated on the left side of the panel.
Analysis of the CRKL and BCR-ABL Interaction in the ΔP1,P2 Mutant
The above data suggested that CRKL was binding to BCR-ABL through another protein or there was another direct binding site for CRKL in full-length BCR-ABL. As the ABL construct that we had used initially in our yeast two-hybrid analysis only contained the SH2, kinase, and a portion of the C-terminus of ABL (Fig 1), it is possible that we missed an interaction with another portion of full-length BCR-ABL. As p185BCR-ABL with a deletion of the P1,P2 region also interacted with CRKL, this suggests that the interaction site is not in the second or third exon of BCR that constitutes the difference between these two molecules.
Three experiments were devised to help in distinguishing between direct and indirect interactions. Cellular lysates of the p210ΔP1,P2 were analyzed for binding to the various GST-CRKL constructs. Compared with full-length BCR-ABL, lysates from the proline-deletion mutant of BCR-ABL had minimal binding of BCR-ABL to the full-length CRKL GST fusion constructs and no binding to the amino terminal SH3 domain of CRKL (Fig 7). The 32Dp210 and 32Dp210ΔP1,P2 lysates were normalized for BCR-ABL expression before this analysis (data not shown). As can be seen from Fig 7, c-ABL from the p210ΔP1,P2 lysate remained capable of being removed from the lysate by the GST-CRKL fusion construct. Although this experiment suggests that the major site of interaction between BCR-ABL and CRKL is removed by the deletion of the proline-rich regions in the C-terminus of ABL, this experiment cannot distinguish direct from indirect interactions.
Binding of BCR-ABL proteins to bacterially expressed CRKL. Lysates from 32D, 32Dp210, or 32Dp210ΔP1,P2 cells were analyzed for binding to (A) GST-SH3n or (B) GST-CRKL. Bound proteins were separated by SDS-PAGE and transferred to PVDF membranes. ABL proteins were detected using the 24-21 ABL MoAb. The 32Dp210 and 32Dp210ΔP1,P2 lysates were normalized for BCR-ABL expression before this analysis. Molecular weight markers are indicated on the left side of the panel.
Binding of BCR-ABL proteins to bacterially expressed CRKL. Lysates from 32D, 32Dp210, or 32Dp210ΔP1,P2 cells were analyzed for binding to (A) GST-SH3n or (B) GST-CRKL. Bound proteins were separated by SDS-PAGE and transferred to PVDF membranes. ABL proteins were detected using the 24-21 ABL MoAb. The 32Dp210 and 32Dp210ΔP1,P2 lysates were normalized for BCR-ABL expression before this analysis. Molecular weight markers are indicated on the left side of the panel.
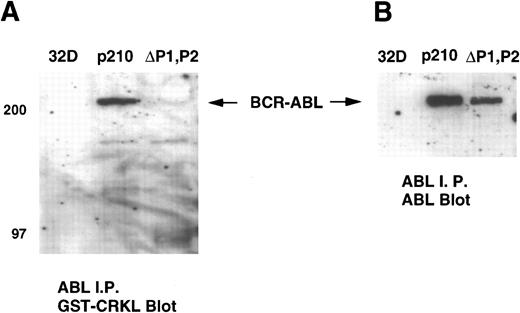
We next performed a gel overlay assay on ABL immunoprecipitates from 32Dp210 and 32Dp210ΔP1,P2 lysates with a GST-CRKL fusion construct. Immunoblots were detected with an anti-GST antisera. These experiments showed that CRKL or CRKL-SH3n bound to full-length BCR-ABL but not the ΔP1,P2 mutant (Fig 8A). Again, similar amounts of BCR-ABL were present in the ABL immunoprecipitates (Fig 8B). No binding to ABL was detected using CRKL-SH2 or GST alone (data not shown). c-ABL was not detected on these immunoblots as the antisera used for these experiments precipitated BCR-ABL more efficiently than c-ABL.
Gel overlay assay of CRKL binding to BCR-ABL. Lysates of 32D, 32Dp210 and 32Dp210ΔP1,P2 cells were immunoprecipitated with ABL antisera K-12, separated by SDS-PAGE, and transferred to PVDF membranes. Immunoblots were probed with (A) GST-CRKL and binding detected with a GST antibody or (B) ABL MoAb 24-21. Molecular weight markers are indicated on the left side of the panel.
Gel overlay assay of CRKL binding to BCR-ABL. Lysates of 32D, 32Dp210 and 32Dp210ΔP1,P2 cells were immunoprecipitated with ABL antisera K-12, separated by SDS-PAGE, and transferred to PVDF membranes. Immunoblots were probed with (A) GST-CRKL and binding detected with a GST antibody or (B) ABL MoAb 24-21. Molecular weight markers are indicated on the left side of the panel.
Finally, the ΔP1,P2 mutant was repackaged into p185BCR-ABL and expressed as a LexA fusion protein for analysis in the yeast two-hybrid system. In this assay, CRKL and p185BCR-ABL were capable of an interaction. However, there was no interaction of CRKL with the ΔP1,P2 mutant (Fig 4).
Cellular Interactions of CRKL
All of the above data suggested that the P1,P2 region mediated a direct interaction of BCR-ABL and CRKL. However, in cells expressing the ΔP1,P2 mutant, CRKL was still tyrosine phosphorylated and associated with BCR-ABL, presumably through another signaling protein. Numerous proteins are known to be tyrosine phosphorylated in BCR-ABL expressing cells and many of them have been found to be capable of forming a complex with BCR-ABL. Further, many of the cellular interactions of CRKII have been determined; however, there is less information about the proteins that interact with CRKL.22 31 Therefore, we analyzed numerous proteins for interaction with CRKL and used the CRKL GST fusion constructs to determine the region of CRKL that mediated binding to these various intracellular proteins.
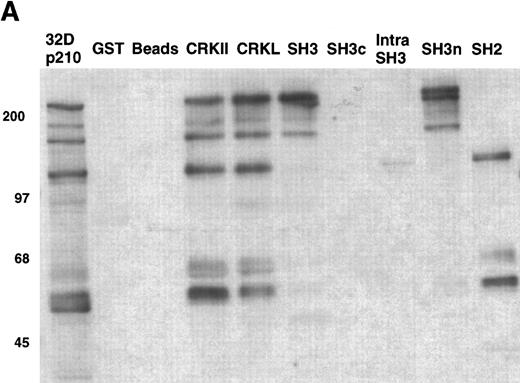
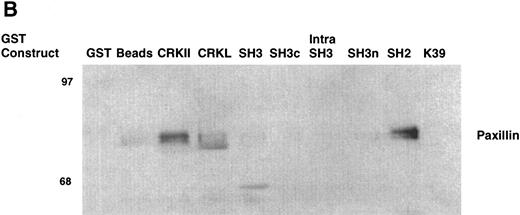
As an example, the pattern of phosphotyrosine proteins that bind to CRKL is shown in Fig 9A. As can be seen, tyrosine phosphorylated proteins of 210, 170, 135-140, 120, 68-70, 62, and 56 kD are capable of binding to full-length CRKL. This is similar to the pattern of tyrosine phosphorylated proteins seen in a CRKL immunoprecipitate (Fig 6A). Interestingly, some of these tyrosine-phosphorylated proteins bind to the amino terminal SH3 domain, particularly p210, p170, and p135-140 while the others bind, as expected, to the SH2 domain. Specific antisera were used to characterize the CRKL interactions. Thus, as previously seen, p210BCR-ABL binds to the amino terminal SH3 domain of CRKL (Fig 3). Other proteins that bind to the amino terminal SH3 domain include p170 SOS, p145 C3G, and p140 c-ABL (Table 1). Proteins that bind to the CRKL SH2 domain include p130 CAS, p120 CBL, and p68 paxillin (Fig 9B and Table 1). No proteins have been identified that bind to the C-terminal SH3 domain of CRKL and the 56- and 62-kD tyrosine-phosphorylated proteins that bind to CRKL have not been identified. Other proteins that were analyzed for binding to CRKL that are known to bind to BCR-ABL, but were negative for binding to CRKL include GRB-2, the 85-kD subunit of phosphatidylinositol 3-kinase, p120 rasGAP, and SHC.
Binding of cellular proteins to bacterially expressed CRKL. Lysates from 1 × 107 32Dp210 cells were bound to GST, glutathione beads (beads), or the various GST-CRKL constructs as described in Materials and Methods. Bound proteins were separated by SDS-PAGE and transferred to PVDF membranes. (A) Tyrosine-phosphorylated proteins were detected with 4G10, a monoclonal antiphosphotyrosine antibody and (B) paxillin was detected with an anti-paxillin MoAb. Molecular weight markers are indicated on the left side of each panel. The lane marked 32Dp210 in (A) is a whole-cell lysate lane.
Binding of cellular proteins to bacterially expressed CRKL. Lysates from 1 × 107 32Dp210 cells were bound to GST, glutathione beads (beads), or the various GST-CRKL constructs as described in Materials and Methods. Bound proteins were separated by SDS-PAGE and transferred to PVDF membranes. (A) Tyrosine-phosphorylated proteins were detected with 4G10, a monoclonal antiphosphotyrosine antibody and (B) paxillin was detected with an anti-paxillin MoAb. Molecular weight markers are indicated on the left side of each panel. The lane marked 32Dp210 in (A) is a whole-cell lysate lane.
Summary of Proteins That Bind to CRKL
| Protein . | Binding Site of CRKL . |
|---|---|
| BCR-ABL | SH3n |
| c-ABL | SH3n |
| C3G (145kD) | SH3n |
| SOS (170 kD) | SH3n |
| *Paxillin (68 kD) | SH2 |
| *CBL (120 kD) | SH2 |
| CAS (130 kD) | SH2 |
| Protein . | Binding Site of CRKL . |
|---|---|
| BCR-ABL | SH3n |
| c-ABL | SH3n |
| C3G (145kD) | SH3n |
| SOS (170 kD) | SH3n |
| *Paxillin (68 kD) | SH2 |
| *CBL (120 kD) | SH2 |
| CAS (130 kD) | SH2 |
Indicates proteins that are also reported to bind to BCR-ABL.
DISCUSSION
In the current study, we have mapped the site of direct interaction of CRKL and BCR-ABL to the amino terminal SH3 domain of CRKL with a proline-rich region in the C-terminus of ABL. Deletion of either proline-rich region abolished binding to CRKL, suggesting that both proline-rich sequences may be required for CRKL binding. Although our initial analysis of binding in the yeast two-hybrid system was performed with a fragment of ABL, we confirmed that there were no other sites of direct binding of CRKL to BCR-ABL by expressing full-length p185BCR-ABL and a p185BCR-ABL with the P1,P2 deletions fused to the DNA binding domain of LexA. This assay showed that there was no binding of CRKL to the p185ΔP1,P2 mutant. Additional evidence for the lack of a direct interaction with the deletion mutant was obtained using a gel overlay assay.
Despite the lack of a direct interaction between BCR-ABL and CRKL in the ΔP1,P2 mutant of BCR-ABL, there was no defect in transforming ability of this mutant as assessed by growth factor independence of myeloid cells and fibroblast transformation. Similar data has been obtained independently by Senechal et al.32 Although our initial impression was that CRKL was not required for BCR-ABL function, analysis of the mutant cell lines showed tyrosine phosphorylation of CRKL and persistent coimmunoprecipitation of CRKL with the proline deletion BCR-ABL mutant. Thus, we cannot exclude a significant role for CRKL in transformation by BCR-ABL via indirect interactions.
Numerous proteins have been shown to interact with BCR-ABL and many have been shown to be tyrosine phosphorylated in BCR-ABL–expressing cells. Besides CRKL, these proteins include rasGAP,33 GRB-2,34,35 SHC,36,37 FES,38 SYP,39 CBL,40-42 the 85-kD subunit of phosphatidylinositol 3-kinase,43 VAV,44 c-BCR,45 paxillin,46 and various other cytoskeletal proteins.47 These proteins and others are candidates for mediating the indirect interaction of CRKL with BCR-ABL. We analyzed the ability of several of these tyrosine-phosphorylated proteins to interact with CRKL and found that p120 CBL and p68 paxillin bind to the SH2 domain of CRKL, while binding to the amino-terminal SH3 domain of CRKL was demonstrated for p210 BCR-ABL. No binding of CRKL to other proteins that are known to associate with BCR-ABL was found, including the 85-kD subunit of PI 3 kinase, GRB-2, SHC, or p120 rasGAP.
Of the proteins that interact with CRKL, paxillin and CBL also interact with BCR-ABL. Either of these proteins or another unidentified protein could be responsible for mediating a ternary complex between BCR-ABL and CRKL in the proline deletion mutant. In the case of CBL, several groups have shown that c-CBL is tyrosine phosphorylated in BCR-ABL–expressing cells and that BCR-ABL and c-CBL are found in a complex as demonstrated by co-immunoprecipitation.40-42 The interaction of c-CBL and BCR-ABL has been shown to be dependent on tyrosine phosphorylation of CBL.42 Additional studies have shown that the ABL SH2 domain is capable of direct binding to tyrosine phosphorylated c-CBL48 and, as shown here, the SH2 domain of CRKL is also capable of binding to c-CBL. Consistent with this, c-CBL immunoprecipitates from BCR-ABL cells not only contain BCR-ABL, but also contain CRKL.48 Thus, our current model is that tyrosine-phosphorylated c-CBL binds to the SH2 domain of ABL in BCR-ABL and, through another tyrosine-phosphorylated residue, binds the SH2 domain of CRKL. The interaction of CRKL and BCR-ABL can also occur directly through the amino terminal domain of CRKL binding to a proline-rich region of ABL.
A prediction of this model would be that BCR-ABL should be capable of binding to the GST-SH2 domain of CRKL by virtue of tyrosine phosphorylated CBL binding to BCR-ABL and the SH2 domain of CRKL. This prediction was confirmed in our GST-fusion protein experiments. It would also be predicted that the GST–full-length CRKL would still be capable of binding some BCR-ABL from the proline deletion, through the interaction of CBL with the SH2 domain of CRKL. Again, this prediction was borne out by the data.
However, it is possible that more complicated complexes occur or that proteins other than CBL mediate an indirect interaction between CRKL and BCR-ABL. For example, the SH2 domain of GRB-2 is known to bind to Tyr 177 of BCR-ABL and its SH3 domain has been shown to bind to CBL.49 As tyrosine-phosphorylated CBL binds to the SH2 domain of CRKL, it is possible that GRB-2 binding to BCR-ABL allows CRKL to associate with BCR-ABL through this interaction with CBL. Given the multitude of complexes induced by BCR-ABL, combinations of mutants (eg, Tyr 177 to Phe with a deletion of the proline-rich sequences in the C-terminus of ABL) will be required to determine the roles of various signaling proteins in BCR-ABL transformation. Alternatively, the ability to BCR-ABL to transform cell lines generated from mice lacking various of these signaling proteins could also be informative.
ACKNOWLEDGMENT
We thank Ravi Salgia and James Griffin for supplying antibodies and also thank Ravi Salgia for his assistance with the gel overlay assays. We are also grateful to Brenda Newman for her help with the yeast two-hybrid assay and to Stan Hollenberg for providing the yeast two-hybrid plasmids.
Supported by National Institutes of Health Grant No. CA65823 and the American Cancer Society, Oregon Division. B.J.D. is a recipient of a Translational Research Award from the Leukemia Society of America.
The first three authors contributed equal amounts of work to this manuscript and should be considered as co-first authors.
Address reprint requests to Brian J. Druker, MD, Division of Hematology and Medical Oncology, L592, Oregon Health Sciences University, 3181 SW Sam Jackson Park Rd, Portland, OR 97201.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal