Abstract
We found a marked variation in BCL-2 oncoprotein expression levels of primary leukemic cells from 338 children with newly diagnosed acute lymphoblastic leukemia (ALL). None of the high-risk features predictive of poor treatment outcome in childhood ALL, such as older age, high white blood cell (WBC) count, organomegaly, T-lineage immunophenotype, ability of leukemic cells to cause overt leukemia in severe combined immunodeficient (SCID) mice, presence of MLL-AF4, and BCR-ABL fusion transcripts were associated with high levels of BCL-2 expression. Overall, high BCL-2 levels were not associated with slow early response, failure to achieve complete remission, or poor event-free survival. High BCL-2 levels in primary leukemic cells predicted slow early response only in T-lineage ALL patients, which comprised approximately 15% of the total patient population. Even for this small subset of patients, the level of BCL-2 expression did not have a significant impact on the short-term event-free survival.
BCL-2 PROTEIN PROMOTES cell survival by preventing apoptosis.1-7 Several studies indicate that the protein expression levels of BCL-2 determine the in vitro sensitivity of lymphoma and leukemia cells to chemotherapeutic drugs.8-14 Initial studies in acute lymphoblastic leukemia (ALL) patients regarding the clinical significance of the interpatient heterogeneity in BCL-2 protein expression did not show a consistent correlation between BCL-2 expression levels in primary leukemic cells and patients' characteristics or treatment outcome.15-17 The purpose of the present Children's Cancer Group (CCG) study was to examine the BCL-2 protein expression levels in primary leukemic cells from a large cohort of 338 children with newly diagnosed ALL, including 286 B-lineage ALL patients and 52 T-lineage ALL patients, in relationship to more commonly measured clinical and laboratory parameters.
MATERIALS AND METHODS
Study patients.Three hundred and thirty eight children with newly diagnosed ALL greater than 1 year of age (ie, noninfants) who were enrolled in three risk-adjusted CCG treatment studies (ie, CCG-1922 for standard risk ALL; CCG-1882 and CCG-1901 for poor risk ALL18-20; from December 1993 through December 1994 were included in this correlative laboratory investigation. There were 140 girls and 198 boys with a median age of 5.3 years (range, 1.0 to 20.7 years). The induction regimens employed vincristine, prednisone (or dexamethasone), L-asparaginase, plus daunomycin in CCG 1882 study; vincristine, prednisone, L-asparaginase, daunomycin plus cyclophosphamide in CCG-1901 study; and vincristine, prednisone plus L-asparaginase in CCG 1922 study. Each protocol was approved by the National Cancer Institute (NCI), as well as the institutional review boards of the CCG-affiliated institutions that participated in this study. Informed consent was obtained from parents, patients, or both, as deemed appropriate, for both treatment and laboratory studies according to guidelines of the Department of Health and Human Services (DHHS) guidelines. The diagnosis was based on morphologic, biochemical, and immunologic features of leukemic cells, including lymphoblast morphology on Wright-Giemsa stained smears of bone marrow, positive nuclear staining for terminal deoxynucleotidyl transferase (TdT), negative staining for myeloperoxidase, and expression of two or more lymphoid differentiation antigens on the plasma membrane (see below). Standard definitions of bone marrow status and early response to therapy were previously reported.18-28 Results were also presented according to the NCI risk classification criteria. These criteria classify a patient as being in the standard risk category if age at diagnosis is 1 to 9 years and the WBC is less than 50,000/μL. Poor risk patients include those who are diagnosed at an age of 10 years or greater and/or who had an initial WBC equal to or greater than 50,000/μL. Patient characteristics are detailed in Table 1. For the Western blot analysis of BCL-2 expression, we used diagnostic primary bone marrow samples under the exemption category (45 CFR Part 46.101;b category 4 Existing data; Records review; Pathologic Specimens) in accordance with DHHS guidelines. These specimens were obtained from routine diagnostic bone marrow aspirates before therapy and informed consent for treatment was obtained from parents, patients, or both based on DHHS guidelines.
Patient Characteristics
| Variable . | No. . | Category . | No. (%) of Evaluable Patients . |
|---|---|---|---|
| Sex | 338 | Female | 140 (41.4) |
| Male | 198 (58.6) | ||
| Age | 338 | <10 yr | 261 (77.2) |
| ≥10 yr | 77 (22.8) | ||
| WBC at diagnosis | 338 | <50 × 109/L | 237 (70.1) |
| ≥50 × 109/L | 101 (29.9) | ||
| NCI risk assignment | 338 | Standard risk | 183 (54.1) |
| Poor risk | 155 (45.9) | ||
| Lymphadenopathy | 338 | No | 166 (49.1) |
| Yes | 172 (50.9) | ||
| Splenomegaly | 337 | No | 158 (46.9) |
| Yes | 179 (53.1) | ||
| Hepatomegaly | 337 | No | 159 (47.2) |
| Yes | 178 (52.8) | ||
| Mediastinal mass | 338 | No | 295 (87.3) |
| Yes | 43 (12.7) | ||
| Immunophenotype | 338 | B-lineage | 286 (84.6) |
| T-lineage | 52 (15.4) | ||
| Molecular genetics | 215 | PCR neg. | 191 (88.8) |
| MLL-AF-4+ [t(4; 11)] | 4 (1.9) | ||
| E2A-PBX-1+ [t(1; 19)] | 11 (5.1) | ||
| BCR-ABL+ [t(9; 22)] | 9 (4.2) | ||
| SCID mouse assay | 295 | No Leukemia | 232 (78.6) |
| Leukemia | 63 (21.4) | ||
| Treatment response | |||
| Day 7 BM status | 324 | M1 or M2 (RER) | 246 (75.9) |
| M3 (SER) | 78 (24.1) | ||
| Day 28 BM status | 336 | M1 (CR) | 329 (97.9) |
| M2 or M3 (no CR) | 7 (2.1) |
| Variable . | No. . | Category . | No. (%) of Evaluable Patients . |
|---|---|---|---|
| Sex | 338 | Female | 140 (41.4) |
| Male | 198 (58.6) | ||
| Age | 338 | <10 yr | 261 (77.2) |
| ≥10 yr | 77 (22.8) | ||
| WBC at diagnosis | 338 | <50 × 109/L | 237 (70.1) |
| ≥50 × 109/L | 101 (29.9) | ||
| NCI risk assignment | 338 | Standard risk | 183 (54.1) |
| Poor risk | 155 (45.9) | ||
| Lymphadenopathy | 338 | No | 166 (49.1) |
| Yes | 172 (50.9) | ||
| Splenomegaly | 337 | No | 158 (46.9) |
| Yes | 179 (53.1) | ||
| Hepatomegaly | 337 | No | 159 (47.2) |
| Yes | 178 (52.8) | ||
| Mediastinal mass | 338 | No | 295 (87.3) |
| Yes | 43 (12.7) | ||
| Immunophenotype | 338 | B-lineage | 286 (84.6) |
| T-lineage | 52 (15.4) | ||
| Molecular genetics | 215 | PCR neg. | 191 (88.8) |
| MLL-AF-4+ [t(4; 11)] | 4 (1.9) | ||
| E2A-PBX-1+ [t(1; 19)] | 11 (5.1) | ||
| BCR-ABL+ [t(9; 22)] | 9 (4.2) | ||
| SCID mouse assay | 295 | No Leukemia | 232 (78.6) |
| Leukemia | 63 (21.4) | ||
| Treatment response | |||
| Day 7 BM status | 324 | M1 or M2 (RER) | 246 (75.9) |
| M3 (SER) | 78 (24.1) | ||
| Day 28 BM status | 336 | M1 (CR) | 329 (97.9) |
| M2 or M3 (no CR) | 7 (2.1) |
Leukemic cells are examined for the presence of MLL-AF-4, E2A-PBX-1 and BCR-ABL by RT-PCR, as described in Materials and Methods.
Abbreviations: BM, bone marrow; WBC, white blood count; BM status: M1, <5% blasts; M2, 5-25% blasts; M3, >25% blasts.
Immunophenotyping.Mononuclear cell fractions containing ≥90% leukemic cells were isolated from pretreatment bone marrow aspirate samples by centrifugation of the cell suspensions on Ficoll-Hypaque gradients. Immunophenotyping was performed centrally in the CCG ALL Biology Reference Laboratory by indirect immunofluorescence and flow cytometry using monoclonal antibodies reactive with the following lymphoid differentiation antigens CD1, CD2, CD3, CD4, CD5, CD7, CD8, CD9, CD10, CD19, CD20, CD21, CD22, CD24, and CD40, as previously described.19,29-35 Cases were classified as B-lineage ALL if ≥30% of leukemic cells were positive for CD19 or CD24 and <30% were positive for CD2, CD5, and CD7.19,30,35 Patients were classified as T-lineage ALL if ≥30% of blasts were positive for CD2, CD5, or CD7 and <30% were positive for CD19 or CD24.19 29
Western blot analysis of BCL-2 protein expression in ALL cells.The expression levels of BCL-2 oncoprotein in whole cell lysates of primary leukemic cells were measured centrally in the Children's Cancer Group ALL Biology Reference Laboratory in Roseville, MN, by Western blot analysis using a monoclonal mouse antihuman BCL-2 antibody (DAKO-bcl–2, Code M857, Lot 063, DAKO, Glostrup, Denmark). In brief, leukemic cells (5 × 106 cells/sample) were lysed in 150 μL sodium dodecyl sulfate (SDS) lysis buffer (50 mmol/L Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 100 μmol/L sodium orthovanadate, 25 mmol/L DTT) and boiled for 5 minutes. The DNA was sheared by several passages through a 28-gauge needle, and 40 μL amounts of the whole cell lysate protein samples in SDS-reducing sample buffer were fractionated on reducing SDS-polyacrylamide gels by overnight electrophoresis at 4 mA. The proteins were transferred to a 0.45-μm Immobilon-polyvinylidene difluoride (PVDF ) membrane (Millipore Corp, Bedford, MA) for 1 hour at 130 mA using a semidry transfer apparatus (Hoefer Scientific Instruments, San Francisco, CA). The PVDF membranes were incubated for 1 hour at room temperature in blocking solution (10 mmol/L Tris-HCl, pH 7.5, 100 mmol/L NaCl, 5% bovine serum albumin), washed in rinsing buffer, and incubated for 1 hour with a mouse monoclonal anti-Bcl–2 antibody (DAKO, Denmark; Code No. M887, Lot 063) at 1 μg/mL final concentration in blocking solution or a mixture of mouse monoclonal anti-Bcl–2 (1 μg/mL) plus mouse monoclonal anti-Actin (Sigma Chemical Co, St Louis, MO; Cat. No A-4700) (1:50,000 final dilution), followed by three 10-minute washes in rinsing buffer. Blots were incubated for 45 minutes with a 125I-labeled sheep antimouse IgG, washed, dried, and autoradiographed by a 24-hour or 48-hour exposure using a Kodak-X-OMAT film (Eastman Kodak Co, Rochester, NY), as previously described.36,37 In some experiments, PVDF blots were stained with Coomassie Blue after autoradiography to exclude formally any nonselective technical problems in transfer of proteins. All autoradiograms were subjected to densitometric scanning using the automated AMBIS system (Automated Microbiology System, Inc, San Diego, CA)38-40 and each case was assigned a unit value for BCL-2 expression by comparing the density of the BCL-2 band to the density of the standard BCL-2 band in the control whole cell lysate sample prepared from 5 × 106 NALM-6 leukemia cells. A total of 100 U of BCL-2 correspond to the amount of BCL-2 protein present in each NALM-6 cell. NALM-6 cell line was used as a control because it is a CD10+ common ALL cell line and its drug sensitivity profile is well established.36 41 In some experiments, instead of the 125I-labeled sheep antimouse IgG and autoradiography, a horseradish peroxidase (HRP)-conjugated sheep antimouse IgG (Signal Transduction Laboratories, Lexington, KY; 1:2,500 dilution, 45-minute incubation) and the ECL chemiluminescence detection system (Amersham Life Sciences, Arlington Heights, IL) were used according to the manufacturers' recommendations.
Detection of fusion transcripts by polymerase chain reaction (PCR).All PCR assays were performed centrally in the CCG Biology Reference Laboratory. Total cellular RNA was extracted from primary leukemic cells patient cells and examined for the presence of t(9; 22)-specific BCR-ABL, t(1; 19)-specific E2A-PBX1, and t(4; 11)-specific MLL/AF-4 fusion transcripts using a reverse-transcriptase (RT) PCR assay, as previously described.42-44 cDNA was synthesized using 20% of the total RNA sample with 100 U of Moloney murine leukemia virus (MMLV) reverse transcriptase (GIBCO-BRL, Gaithersburg, MD) in 1× PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-Cl pH 8.3, 1.5 mmol/L MgCl2 , and 0.01% (wt/vol) gelatin), 3 μg random primers (GIBCO-BRL), 2.5 mmol/L of each of the deoxyribonucleoside triphosphates (dNTPs) (Pharmacia, Piscataway, NJ), 0.01 mol/L DTT, and 20 U RNasin (Promega, Madison, WI) in a total volume of 20 μL at 37°C for 1 hour, as described.42-44 The reaction mixture (Reaction mixture 1) was then denatured at 95°C for 5 minutes and cooled on ice. For amplification of the cDNA products, samples were diluted fivefold with 1× PCR buffer containing 0.075 μg of each oligonucleotide primer, 6.25% dimethyl sulfoxide (DMSO) and 2.5 U Amplitaq DNA Polymerase (Perkin Elmer Cetus, Norwalk, CT) (Reaction mixture 2) and 35 cycles of amplification were performed with a DNA thermal cycler, as reported.42-44 Primers used were as follows: 5′ TCCGAGGCCACCATCGTGGGCGTCCGC-3′ and 5′-TGTGATTATAGCCTAAGACCCGGAG-3′ for BCR/ABL; 5′-GCCAGCCAGGCACCCTCCC-3′ and 5′-GTTGTCCAGCCGCATCAGCT-3′ for E2A/PBX1, and 5′-AGAGCAGAGCAAACAGAA-3′ and 5′-GCTGAGAATTTGAGTGAG-3′ for MLL/AF-4. RNA integrity was analyzed on every sample using an ABL RT-PCR with primers 5′-TTCAGCGGCCAGTAGCATCTGACTT-3′ and 5′-TGTGATTATAGCCTAAGACCCGGAG-3′. PCR products were then separated by electrophoresis in a 1.2% agarose gel, transferred to nylon membranes by the method of Southern, and hybridized with previously reported oligonucleotide probes, which were 5′ end-labeled with γ 32P-adenosine triphosphate (ATP) (ICN Pharmaceuticals, Plainview, NY) and T4 Polynucleotide Kinase (Promega), as described.42-44 Autoradiography was performed with Kodak XAR-5 film. Controls included PCR reaction products of RNA-free reaction mixture 1 plus reaction mixture 2 (= Negative control [1]), RNA-free reaction mixture 2 (=Negative control [2]), as well as PCR products from RNA samples of two ALL patients with known absence of BCR-ABL, E2A-PBX1, and MLL-AF4 fusion transcripts (=Negative control [3] and Negative control [4]).
Interpatient variation in BCL-2 oncoprotein expression levels of primary leukemic cells from children with newly diagnosed childhood ALL. In two independent experiments depicted in (A)] and (B), which yielded virtually identical results, BCL-2 and actin expression levels of leukemic cells from 11 patients were compared by Western blot analysis using the ECL chemiluminiscence detection system, as described in Materials and Methods. In (A), the BCL-2 levels ranged from 14 U/blast (pt. no. 3, BCL-2/actin ratio = 0.10) to 160 U/blast (pt. no. 2, BCL-2/actin ratio = 1.13). In (B), duplicate samples from the same patients were subjected to an independent Western blot analysis. Consistent with the initial results shown in (A), the BCL-2 levels ranged from 14 U/blast in pt. no. 3 (BCL-2/actin ratio = 0.15) to 173 U/blast in pt. no. 2 (BCL-2/actin ratio = 1.15). NALM-6 cell lysates were included in each experiment as an internal standard to assign a unit value to the BCL-2 expression levels of primary leukemic cells. Arrow indicates the position of the 27 kD BCL-2 protein.
Interpatient variation in BCL-2 oncoprotein expression levels of primary leukemic cells from children with newly diagnosed childhood ALL. In two independent experiments depicted in (A)] and (B), which yielded virtually identical results, BCL-2 and actin expression levels of leukemic cells from 11 patients were compared by Western blot analysis using the ECL chemiluminiscence detection system, as described in Materials and Methods. In (A), the BCL-2 levels ranged from 14 U/blast (pt. no. 3, BCL-2/actin ratio = 0.10) to 160 U/blast (pt. no. 2, BCL-2/actin ratio = 1.13). In (B), duplicate samples from the same patients were subjected to an independent Western blot analysis. Consistent with the initial results shown in (A), the BCL-2 levels ranged from 14 U/blast in pt. no. 3 (BCL-2/actin ratio = 0.15) to 173 U/blast in pt. no. 2 (BCL-2/actin ratio = 1.15). NALM-6 cell lysates were included in each experiment as an internal standard to assign a unit value to the BCL-2 expression levels of primary leukemic cells. Arrow indicates the position of the 27 kD BCL-2 protein.
Expression of BCL-2 Oncoprotein in Leukemic Cells From Children With Newly Diagnosed ALL
| Patient Subset . | N . | BCL-2 Level (U/blast) . | P Value . | ||
|---|---|---|---|---|---|
| . | . | Mean (SE) . | Median . | Range . | . |
| All | 338 | 67.6 (2.4) | 65.0 | 0.0-221.0 | — |
| Female | 140 | 65.6 (3.7) | 64.5 | 0.0-210.0 | |
| Male | 198 | 68.7 (3.1) | 65.0 | 0.0-221.0 | NS |
| Age <10 yr | 261 | 68.6 (2.8) | 68.0 | 0.0-221.0 | |
| Age ≥10 yr | 77 | 63.4 (4.8) | 59.0 | 0.0-196.0 | NS |
| WBC <50 × 109/L | 237 | 71.6 (2.8) | 70.0 | 0.0-221.0 | |
| WBC ≥50 ×109/L | 101 | 57.7 (4.3) | 44.0 | 0.0-180.0 | .008 |
| NCI standard risk | 183 | 71.7 (3.2) | 73.0 | 1.0-221.0 | |
| NCI poor risk | 155 | 62.4 (3.6) | 55.0 | 0.0-196.0 | .053 |
| Without lymphadenopathy | 166 | 71.6 (3.5) | 73.5 | 0.0-210.0 | |
| With lymphadenopathy | 172 | 63.4 (3.3) | 59.0 | 0.0-221.0 | .088 |
| Without splenomegaly | 158 | 75.1 (3.8) | 72.0 | 0.0-221.0 | |
| With splenomegaly | 179 | 60.5 (3.0) | 56.0 | 0.0-171.0 | .001 |
| Without hepatomegaly | 159 | 72.4 (3.6) | 70.0 | 0.0-210.0 | |
| With hepatomegaly | 178 | 62.9 (3.1) | 59.8 | 0.0-221.0 | .049 |
| Without mediastinal mass | 295 | 69.9 (2.5) | 70.0 | 0.0-221.0 | |
| With mediastinal mass | 43 | 50.4 (6.7) | 41.0 | 0.0-196.0 | .006 |
| B-lineage ALL | 286 | 72.5 (2.6) | 73.0 | 0.0-221.0 | |
| T-lineage ALL | 52 | 40.3 (5.0) | 33.3 | 0.0-196.0 | <.001 |
| PCR-ALL | 191 | 70.2 (3.2) | 66.8 | 0.0-210.0 | |
| MLL-AF-4+ ALL | 4 | 61.4 (20.5) | 64.0 | 16.0-101.5 | NS |
| E2A-PBX-1+ ALL | 11 | 37.3 (8.5) | 24.0 | 9.0-89.0 | .022 |
| BCR-ABL+ ALL | 10 | 56.7 (13.9) | 50.5 | 5.0-125.0 | NS |
| No leukemic cell growth in SCID mice | 232 | 70.1 (2.7) | 69.5 | 0.0-200.0 | |
| Leukemic cell growth in SCID mice | 63 | 57.4 (5.4) | 46.0 | 4.0-210.0 | .035 |
| RER (day 7 BM) | 246 | 68.1 (2.4) | 67.0 | 0.0-221.0 | |
| SER (day 7 BM) | 78 | 66.9 (4.9) | 60.8 | 0.0-200.0 | NS |
| T-lineage ALL, RER | 35 | 32.8 (4.8) | 25.5 | 0.0-102.0 | |
| T-lineage ALL, SER | 14 | 61.9 (12.7) | 59.3 | 0.0-196.0 | .011 |
| B-lineage ALL, RER | 211 | 73.9 (2.5) | 72.0 | 0.0-221.0 | |
| B-lineage ALL, SER | 64 | 68.0 (2.4) | 63.0 | 0.0-200.0 | NS |
| CR (day 28 BM) | 329 | 67.6 (2.4) | 65.0 | 0.0-221.0 | |
| No CR (day 28 BM) | 7 | 72.7 (8.2) | 65.0 | 54.0-117.0 | NS |
| Patient Subset . | N . | BCL-2 Level (U/blast) . | P Value . | ||
|---|---|---|---|---|---|
| . | . | Mean (SE) . | Median . | Range . | . |
| All | 338 | 67.6 (2.4) | 65.0 | 0.0-221.0 | — |
| Female | 140 | 65.6 (3.7) | 64.5 | 0.0-210.0 | |
| Male | 198 | 68.7 (3.1) | 65.0 | 0.0-221.0 | NS |
| Age <10 yr | 261 | 68.6 (2.8) | 68.0 | 0.0-221.0 | |
| Age ≥10 yr | 77 | 63.4 (4.8) | 59.0 | 0.0-196.0 | NS |
| WBC <50 × 109/L | 237 | 71.6 (2.8) | 70.0 | 0.0-221.0 | |
| WBC ≥50 ×109/L | 101 | 57.7 (4.3) | 44.0 | 0.0-180.0 | .008 |
| NCI standard risk | 183 | 71.7 (3.2) | 73.0 | 1.0-221.0 | |
| NCI poor risk | 155 | 62.4 (3.6) | 55.0 | 0.0-196.0 | .053 |
| Without lymphadenopathy | 166 | 71.6 (3.5) | 73.5 | 0.0-210.0 | |
| With lymphadenopathy | 172 | 63.4 (3.3) | 59.0 | 0.0-221.0 | .088 |
| Without splenomegaly | 158 | 75.1 (3.8) | 72.0 | 0.0-221.0 | |
| With splenomegaly | 179 | 60.5 (3.0) | 56.0 | 0.0-171.0 | .001 |
| Without hepatomegaly | 159 | 72.4 (3.6) | 70.0 | 0.0-210.0 | |
| With hepatomegaly | 178 | 62.9 (3.1) | 59.8 | 0.0-221.0 | .049 |
| Without mediastinal mass | 295 | 69.9 (2.5) | 70.0 | 0.0-221.0 | |
| With mediastinal mass | 43 | 50.4 (6.7) | 41.0 | 0.0-196.0 | .006 |
| B-lineage ALL | 286 | 72.5 (2.6) | 73.0 | 0.0-221.0 | |
| T-lineage ALL | 52 | 40.3 (5.0) | 33.3 | 0.0-196.0 | <.001 |
| PCR-ALL | 191 | 70.2 (3.2) | 66.8 | 0.0-210.0 | |
| MLL-AF-4+ ALL | 4 | 61.4 (20.5) | 64.0 | 16.0-101.5 | NS |
| E2A-PBX-1+ ALL | 11 | 37.3 (8.5) | 24.0 | 9.0-89.0 | .022 |
| BCR-ABL+ ALL | 10 | 56.7 (13.9) | 50.5 | 5.0-125.0 | NS |
| No leukemic cell growth in SCID mice | 232 | 70.1 (2.7) | 69.5 | 0.0-200.0 | |
| Leukemic cell growth in SCID mice | 63 | 57.4 (5.4) | 46.0 | 4.0-210.0 | .035 |
| RER (day 7 BM) | 246 | 68.1 (2.4) | 67.0 | 0.0-221.0 | |
| SER (day 7 BM) | 78 | 66.9 (4.9) | 60.8 | 0.0-200.0 | NS |
| T-lineage ALL, RER | 35 | 32.8 (4.8) | 25.5 | 0.0-102.0 | |
| T-lineage ALL, SER | 14 | 61.9 (12.7) | 59.3 | 0.0-196.0 | .011 |
| B-lineage ALL, RER | 211 | 73.9 (2.5) | 72.0 | 0.0-221.0 | |
| B-lineage ALL, SER | 64 | 68.0 (2.4) | 63.0 | 0.0-200.0 | NS |
| CR (day 28 BM) | 329 | 67.6 (2.4) | 65.0 | 0.0-221.0 | |
| No CR (day 28 BM) | 7 | 72.7 (8.2) | 65.0 | 54.0-117.0 | NS |
Abbreviation: NS, not significant.
SCID mouse assays.Primary leukemic cells were examined for their ability to cause leukemia in SCID mice, as previously described.41,43,44 All SCID mice were produced by specific pathogen-free (SPF ) CB-17 scid/scid breeders and maintained in an SPF environment in Microisolator cages (Lab Products, Inc, Maywood, NY). Mice were inoculated with 5 × 106 leukemic cells via tail vein injections. Mice were killed at 12 weeks or when they became moribund as a result of disseminated leukemia. Mice were necropsied at the time of death or euthanization, and their tissues were histopathologically examined for the presence of leukemia cells, as previously reported.41,43 44 For each mouse, multiple tissues, including brain, lung, heart, thymus, liver, kidney, spleen, gut, pancreas, ovary, sagittal sections of a femur, and several vertebrae were histologically evaluated. All histopathologic studies of SCID mouse tissues were performed by a veterinary pathologist in a prospective fashion without any knowledge of BCL-2 expression levels.
Statistical methods.We used standard statistical methods, including Student's t-tests, to evaluate the BCL-2 expression levels in primary ALL cells and to compare the BCL-2 levels of leukemic cells from different patients, as previously described.19,29,30,31,35,41 Possible associations of BCL-2 expression level in primary leukemic cells with clinical, demographic, and laboratory features of the patients from whom cells were derived were examined by using chi-square analyses of categorical groupings. BCL-2 levels of leukemic cells were also correlated on a continuous scale with numerical variables (eg, age, WBC, day 7 bone marrow blast percentage). Event-free survival (EFS) was defined as the time from study entry to the first major event (failure to achieve remission, relapse at any site, death, or second malignancy). For patients who did not experience an event, EFS was defined as the time to last follow-up and treated as a censored life table observation. Estimates of EFS were based on the Kaplan-Meier life table method for censored data.19,29 41 Comparisons of outcome between patient subsets were performed by the log rank test. All computations were performed using an updated statistics program for the Macintosh (StatWorks; Cricket Software, Philadelphia, PA).
RESULTS
The BCL-2 protein expression levels were determined in 338 cases by Western blot analysis using a monoclonal mouse antihuman BCL-2 antibody. As illustrated in Fig 1 and detailed in Table 2, there was a marked interpatient heterogeneity in expression levels of BCL-2 protein, ranging from <0.1 U/cell to 221 U/cell (mean ± standard error [SE] = 67.6 ± 2.4 U/cell; median, 65.0 U/blast). Comparison of the expression levels of the housekeeping protein Actin by Western blot analysis demonstrated that these interpatient differences in Bcl-2 expression were not due to differences in overall protein expression levels or protein loading errors during electrophoresis (Fig 1). When we examined the possible association of BCL-2 protein expression levels in primary leukemic cells with ALL patients' clinical, demographic, and laboratory features, we found that neither age ≥10 years, WBC ≥ 50 × 109/L nor the presence of MLL-AF4 and BCR-ABL fusion transcripts, which are predictive of poor treatment outcome,42,45-49 were associated with high levels of BCL-2 expression. On the contrary, leukemic cells from patients with WBC < 50 × 109/L exhibited significantly higher BCL-2 levels than did leukemic cells from patients with WBC ≥ 50 × 109/L (71.6 ± 2.8 U/cell v 57.7 ± 4.3 U/cell, P = .008). Leukemic cells from patients with standard risk ALL according to the NCI Risk Criteria45 had significantly higher BCL-2 levels than those from poor risk ALL patients (71.7 ± 3.2 U/blast v 62.4 ± 3.6, P = .053; Table 2). Leukemic cells from patients who presented with splenomegaly or hepatomegaly expressed significantly lower levels of BCL-2 than cells from patients without organomegaly (Table 2). The presence of E2A-PBX1 fusion transcript, which has been correlated with poor risk features,45 was also associated with lower levels of BCL-2 protein (P = .022). Leukemic cells from T-lineage ALL patients, who usually present with poor risk features,19,29,46,47 49 expressed 1.8-fold lower levels of BCL-2 protein than leukemic cells from B-lineage ALL patients (40.3 ± 5.0 U/cell v 72.5 ± 2.6 U/cell, P < .001; Table 2).
A recent study demonstrated that growth of primary leukemic cells in SCID mice is a strong and independent predictor of relapse in patients with newly diagnosed high-risk ALL.41 Therefore, we examined whether primary leukemic cells that caused leukemia in SCID mice had higher levels of BCL-2 than leukemic cells that did not grow in SCID mice. Contrary to our initial expectations, leukemic cells that caused leukemia in SCID mice had significantly lower amounts of BCL-2 protein than leukemic cells that did not grow in SCID mice (57.4 ± 5.4 U/cell v 70.1 ± 2.7 U/cell, P = .035) (Table 2).
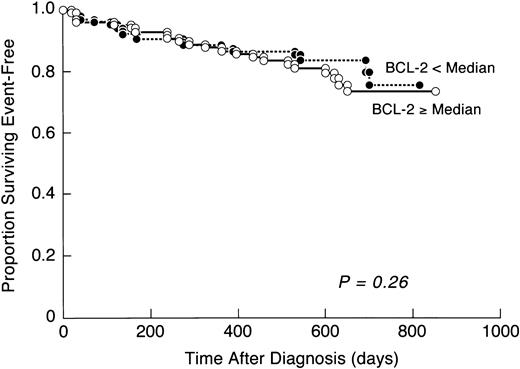
Early response to therapy, which refers to the degree of disease regression before day 28 of induction therapy (viz., the time at which bone marrow remission is conventionally documented) is a highly significant and independent prognostic factor in childhood ALL.18,21,22,45,50-52 Slow early response (SER; defined as M3 marrow status on day 7) implies a greater risk of early and late relapses even when patient strata with relatively homogenous prognostic features are examined.18,21,22,45,50-52 Persistence of >5% leukemic cells in bone marrow by day 28 of induction therapy (that is no complete remission [CR]) is also an indicator of poor outcome.23 As shown in Table 2, leukemic cells from patients with SER did not express higher levels of BCL-2 than did leukemic cells from patients with rapid early response (RER; defined as M1 or M2 marrow status on day 7) (SER v RER: 66.9 ± 4.9 v 68.1 ± 2.8). Similarly, patients who failed to achieve a CR by day 28 did not express high levels of BCL-2. Patients whose leukemic cell BCL-2 content was ≥65 U/cell (=median value, see Table 2), did not appear to have a worse EFS outcome within the first 2 years of study entry than patients whose leukemic cell BCL-2 content was <65 U/cell (Fig 2).
EFS according to BCL-2 expression levels in primary leukemic cells. The cumulative proportions of patients surviving event-free are shown according to levels of BCL-2 expression on leukemic cells. Patients were divided into two groups: BCL-2 level ≥ median (ie, ≥65 U/cell) and BCL-2 level < median (ie, <65 U/cell). BCL-2 level showed no prognostic effect on EFS.
EFS according to BCL-2 expression levels in primary leukemic cells. The cumulative proportions of patients surviving event-free are shown according to levels of BCL-2 expression on leukemic cells. Patients were divided into two groups: BCL-2 level ≥ median (ie, ≥65 U/cell) and BCL-2 level < median (ie, <65 U/cell). BCL-2 level showed no prognostic effect on EFS.
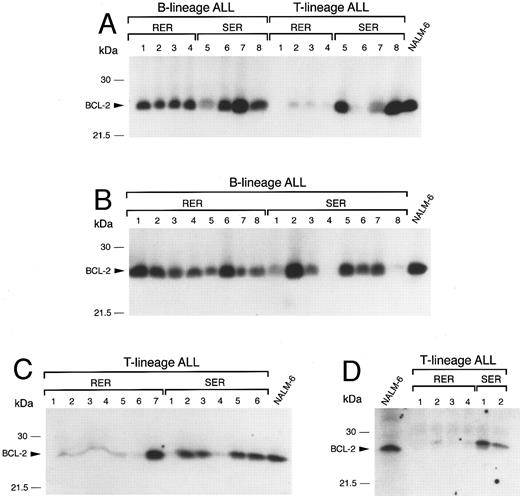
Expression of BCL-2 in immunophenotypically distinct patient subsets according to treatment response. (A) BCL-2 expression in primary leukemic cells from eight B-lineage ALL and eight T-lineage ALL patients. In each subset, one half of the patients were rapid early responders (RER) and the other half slow early responders (SER). (B) BCL-2 expression in primary leukemic cells from 16 B-lineage ALL patients (eight RER and eight SER). (C) + (D) BCL-2 expression in 19 T-lineage ALL patients (11 RER, eight SER). In (A) through (D), NALM-6 cell lysates were used as internal standards.
Expression of BCL-2 in immunophenotypically distinct patient subsets according to treatment response. (A) BCL-2 expression in primary leukemic cells from eight B-lineage ALL and eight T-lineage ALL patients. In each subset, one half of the patients were rapid early responders (RER) and the other half slow early responders (SER). (B) BCL-2 expression in primary leukemic cells from 16 B-lineage ALL patients (eight RER and eight SER). (C) + (D) BCL-2 expression in 19 T-lineage ALL patients (11 RER, eight SER). In (A) through (D), NALM-6 cell lysates were used as internal standards.
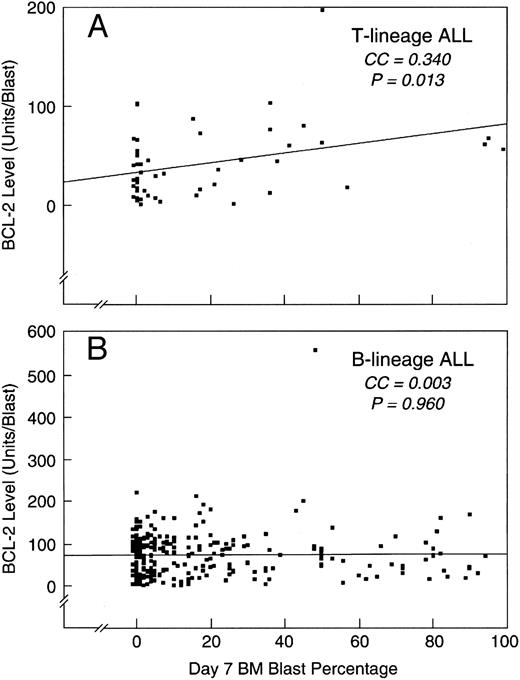
Correlation of BCL-2 levels with the percentage of leukemic cells in the day 7 bone marrow samples. The BCL-2 expression levels were correlated on a continuous scale with the day 7 bone marrow blast percentage by simple regression. (A) T-lineage ALL subset. The coefficient of correlation (CC) was 0.340, with a P = .013. (B) B-lineage ALL subset. The CC was 0.003, with a P = .960.
Correlation of BCL-2 levels with the percentage of leukemic cells in the day 7 bone marrow samples. The BCL-2 expression levels were correlated on a continuous scale with the day 7 bone marrow blast percentage by simple regression. (A) T-lineage ALL subset. The coefficient of correlation (CC) was 0.340, with a P = .013. (B) B-lineage ALL subset. The CC was 0.003, with a P = .960.
When we examined the potential role of BCL-2 expression level in predicting early response to therapy in T-lineage ALL patients separately, we found that leukemic cells from patients with SER had significantly higher BCL-2 levels than those from patients with RER (61.9 ± 12.7 v 32.8 ± 4.8, P = .011, Table 2; Fig 3). The coefficient of correlation for BCL-2 levels and the percentages of residual leukemic cells in day 7 bone marrow samples of T-lineage ALL patients was 0.340, with a P value for statistical significance of .013 (Fig 4A). In contrast, there was no significant correlation between BCL-2 levels and the percentages of residual leukemic cells in day 7 bone marrow samples of B-lineage ALL patients (coefficient of correlation = 0.003, P = .960; Fig 4B) and no differences in BCL-2 expression were discernible between the SER versus RER groups in this subset (Table 2, Fig 3). The level of BCL-2 expression did not have a significant impact on the short-term (ie, within the first 2 years after diagnosis) EFS outcome of T-lineage ALL or B-lineage ALL patients (Fig 5).
EFS according to BCL-2 expression levels in primary leukemic cells in immunophenotypically distinct patient subsets. The cumulative proportions of T-lineage ALL (shown in [A]) and B-lineage ALL (shown in [B]) patients surviving event-free are shown according to levels of BCL-2 expression on leukemic cells. Patients were divided into two groups: BCL-2 level ≥ median and BCL-2 level < median. BCL-2 level showed no prognostic effect on EFS in either subset.
EFS according to BCL-2 expression levels in primary leukemic cells in immunophenotypically distinct patient subsets. The cumulative proportions of T-lineage ALL (shown in [A]) and B-lineage ALL (shown in [B]) patients surviving event-free are shown according to levels of BCL-2 expression on leukemic cells. Patients were divided into two groups: BCL-2 level ≥ median and BCL-2 level < median. BCL-2 level showed no prognostic effect on EFS in either subset.
DISCUSSION
BCL-2 is expressed at high levels in non-Hodgkin's lymphoma (NHL), especially in follicular lymphoma and impedes apoptosis mediated by several agents.53,54 Antisense-mediated downregulation of BCL-2 protein expression in NHL cell lines results in reduced clonogenicity55 and enhanced sensitivity to anticancer drugs.56 Furthermore, antisense oligonucleotides to the bcl-2 gene effectively suppressed in vivo growth of xenografted human NHL cells in SCID-hu mouse models.57 In agreement with the role of BCL-2 in chemotherapy-induced apoptosis, BCL-2 protein expression in NHL was correlated with advanced stage and poor treatment outcome.21,53 Patients with high BCL-2 expression levels have significantly reduced EFS and overall survival than patients with low or intermediate levels of BCL-2 expression.53
Variable levels of BCL-2 protein are expressed in acute myeloid leukemia (AML) cells.58,59 Inhibition of BCL-2 protein expression by bcl-2 antisense oligodeoxynucleotides decreased the life span of AML cells in vitro and augmented their sensitivity to chemotherapeutic agents.60 In one study, high BCL-2 expression levels were correlated with autocrine granulocyte-macrophage colony-stimulating factor (GM-CSF ) production, which has been associated with poor treatment outcome.59 High expression levels of BCL-2 were recently reported to be associated with poor prognostic features and a poor treatment outcome in AML.2,58 61
BCL-2 expression in ALL cells has been investigated in a number of studies.2,12,13,15,17 In one study, the expression levels of BCL-2 protein in primary leukemic cells from ALL patients correlated with their ability to survive in vitro in the absence of stromal support.12 In other studies, a human ALL cell line transfected with bcl-2 gene exhibited significantly prolonged survival and reduced apoptosis in the presence of several chemotherapeutic agents, including vincristine and dexamethasone, which are important components of standard induction chemotherapy regimens.13 However, pilot correlative clinical/laboratory studies in ALL yielded conflicting results regarding the correlation between BCL-2 expression levels and other patient characteristics or treatment outcome.2,15-17 In a recently published single institution study involving 43 children with newly diagnosed B-lineage ALL and nine children with T-lineage ALL, investigators from the St. Jude Children's Hospital found no correlation between cellular levels of BCL-2 protein and in vitro drug sensitivity of leukemic cells or treatment outcome of patients.17
In the present analysis of 338 children with newly diagnosed ALL, we found a profound interpatient heterogeneity in cellular expression levels of BCL-2 oncoprotein. Overall, high BCL-2 levels were not associated with poor risk features at presentation, slow early response, failure to achieve CR, or poor EFS. On the contrary, clinical and laboratory features, which have been commonly associated with poor treatment outcome, such as high WBC, organomegaly, T-lineage immunophenotype, ability of patients' leukemic cells to cause disseminated human leukemia in SCID mice were correlated with significantly lower BCL-2 expression levels. High BCL-2 levels in primary leukemic cells predicted slow early response in T-lineage ALL patients, which comprised approximately 15% of the total patient population. However, with limited follow-up and an excellent overall outcome for T-lineage ALL patients,19 this relationship did not extend to EFS. Many comparisons are presented and further study of the T-lineage ALL patients would seem warranted. Our findings are consistent with and extend previous studies on the clinical relevance of BCL-2 expression in childhood ALL.16 17
For 20% of the children with standard risk ALL and 40% of children with poor risk ALL, contemporary risk-adjusted treatment programs are inadequate.45-48 Because of the high frequency of ALL among childhood malignancies,62,63 treatment failures in ALL make a major contribution to the overall morbidity and mortality of childhood cancer. The predictive power of the early response and remission induction data18,21-23,45,50-52 indicates that inherent resistance of leukemic cells to components of induction chemotherapy significantly contributes to treatment failure in childhood ALL. Proteins involved in triggering or prevention of drug-induced apoptosis are expected to be the key determinants of early response after induction chemotherapy. In view of the documented lack of correlation between BCL-2 expression levels and early response, it would seem highly unlikely that BCL-2 protein plays a pivotal role in resistance to components of the standard induction chemotherapy regimens. It is important to note that BCL-2 is now known to be but one member of a large family of genes that regulate cell survival and drug resistance.15 Future studies of this gene family as well as unrelated genes implicated in apoptotic pathways in relationship to early response and other prognostic factors may help us identify new molecular targets for innovative treatment strategies.
Supported in part by research grants including CCG Chairman's Grant No. CA-13539, CA-51425, CA-42633, CA-42111, CA-60437, and CA-27137 from the National Cancer Institute, National Institutes of Health, Bethesda, MD. F.M.U. is a Stohlman Scholar of the Leukemia Society of America (New York, NY).
Address reprint requests to Fatih M. Uckun, MD, Children's Cancer Group ALL Biology Reference Laboratory, Biotherapy Institute, University of Minnesota Academic Health Center, 2625 Patton Rd, Roseville, MN 55113.

![Fig. 1. Interpatient variation in BCL-2 oncoprotein expression levels of primary leukemic cells from children with newly diagnosed childhood ALL. In two independent experiments depicted in (A)] and (B), which yielded virtually identical results, BCL-2 and actin expression levels of leukemic cells from 11 patients were compared by Western blot analysis using the ECL chemiluminiscence detection system, as described in Materials and Methods. In (A), the BCL-2 levels ranged from 14 U/blast (pt. no. 3, BCL-2/actin ratio = 0.10) to 160 U/blast (pt. no. 2, BCL-2/actin ratio = 1.13). In (B), duplicate samples from the same patients were subjected to an independent Western blot analysis. Consistent with the initial results shown in (A), the BCL-2 levels ranged from 14 U/blast in pt. no. 3 (BCL-2/actin ratio = 0.15) to 173 U/blast in pt. no. 2 (BCL-2/actin ratio = 1.15). NALM-6 cell lysates were included in each experiment as an internal standard to assign a unit value to the BCL-2 expression levels of primary leukemic cells. Arrow indicates the position of the 27 kD BCL-2 protein.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3769/3/m_bl_0038f1.jpeg?Expires=1765050825&Signature=BvFBy2UE2S4XEAuVZ38o9hQl5d57l3VnI9tkka~9~-aONDwQz7G~cuKALleDHhZ7YNjTweJyrynV7XhlT4NE2T11pa-7gvQcwMaMAjARpSpMDb1QTksm4aJxI6JsGNlACfdtdKx9BvsfScf1qCkj5n~Z-Ffzxc~GYiMpZOa6NE1PS42RUlfKEz7HSsPrkOhy55AYVxXb-EPOXri1BXBHOZteIZVmVWPozzF9ZqBA8WkbovZre51AM-mBUjPcn5QURAS2GI60N~obr6lIDSaPZQcGpAGNgguuyMA-PzNNojtnc1xyilg~2kv2UWZDKjnp0rARJAmCVrvNIZAISKWIGA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. EFS according to BCL-2 expression levels in primary leukemic cells in immunophenotypically distinct patient subsets. The cumulative proportions of T-lineage ALL (shown in [A]) and B-lineage ALL (shown in [B]) patients surviving event-free are shown according to levels of BCL-2 expression on leukemic cells. Patients were divided into two groups: BCL-2 level ≥ median and BCL-2 level < median. BCL-2 level showed no prognostic effect on EFS in either subset.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/10/10.1182_blood.v89.10.3769/3/m_bl_0038f5.jpeg?Expires=1765050825&Signature=16m~Xw~Mlk5hRdO2mBayMpQzSV3XqGXarBxSQXBd6dbp0VIF~ujskNomDSldKHVWCpVEDODoIn0I59xzwHf~v94qqia~6dX-vgVNuO4yxQHMBWuBmchiTfyzNTKwr4ZxdWiAzaYOjWALod-qPLNh3pwwzRvf1wZNW7EH3eRDk06PFE49NrYB4zeW65NLbQGQ9W2~TxfKdECdC0v89WzlkDsR2UckcTOmOghBgjviDrwgFk1uWP5MqcIsV0gsSNslgwgwuFlpaI3jTHyuzk~hDQ40hIQOkZnf2KyR0FHa9VcYbfqBYSmkdEsmF2~NEwr-UONSGukUewmtfvFiDcOZ0w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal