Abstract
Hepatitis G virus (HGV) is a newly described virus that has been implicated in transfusion-associated hepatitis. The prevalence of HGV in a group of multitransfused patients with hematological malignancy was studied using a reverse transcription polymerase chain reaction technique. Transfusion histories and serum aspartate aminotransferase (AST) levels were recorded. HGV was detected in 29 of 60 (48%) patients. There was no difference in HGV positivity rates between those with normal AST levels and those with raised AST levels. Analysis of patients by treatment type showed that 20 of 33 (61%) patients who received a bone marrow transplantation procedure were HGV positive compared with 9 of 27 (33%) treated with conventional combination chemotherapy (P = .036) despite similar transfusion histories. There was no significant difference in HGV positivity between patients treated before the introduction of United Kingdom blood donor screening for hepatitis C virus antibody:18 of 39 (46%) and those treated after the introduction of screening 11 of 21 (52%). HGV infection appears to be extremely common in these patients; however, the clinical significance of these findings with respect to liver dysfunction is not yet clear.
A NEW HEPATITIS virus has recently been described by two groups of workers. Simons et al1 discovered two viruses (termed GBV-A and GBV-B) in plasma from tamarins infected with the GB hepatitis agent. This agent originated from the serum of a 34-year-old surgeon (GB) who had acute non–A-E hepatitis. Subsequently, using a reverse transcription polymerase chain reaction (RT-PCR) with degenerate primers they found virus-like nucleotide sequences in the serum of several patients with non–A-E hepatitis, which was reactive in GBV-A–specific enzyme linked immunosorbent assays (ELISA).2 This new virus had a high degree of identity with GBV-A and was named GBV-C. It is an additional member of the Flaviviridae family and is similar to the hepatitis C virus (HCV). Independently, Linnen et al3 identified a virus in the plasma of a patient with chronic hepatitis, which they named hepatitis G virus (HGV). They showed that the virus is associated with acute and chronic hepatitis and that it is transmissible by transfusion. Analysis of the polyprotein sequences of HGV and GBV-C show amino acid sequence identity at 95%, and the two agents are now considered to be two isolates of the same virus.4 For reasons of clarity the virus will be referred to as HGV throughout this report.
It is known that patients who receive multiple transfusions of blood components are at risk of developing post-transfusion hepatitis. Over 90% of cases of non-A non-B transfusion-related hepatitis have been shown to be due to HCV infection.5 In developed countries blood donors are now routinely screened for anti-HCV with the aim of reducing transfusion-related viral hepatitis. Because the HGV virus may be transmitted by blood transfusion,3,6 7 we have examined sera, using an RT-PCR technique, from a group of patients who have received multiple blood component transfusions during their treatment for hematologic malignancy. Our aims were to establish the prevalence of HGV in these patients and to make some assessment of its clinical relevance.
In one study, 18% of 109 patients with acute HCV were also HGV-RNA positive.3 In view of this, we have also tested our patients for HCV and assessed if the introduction of blood donor screening for anti-HCV in the United Kingdom has had a concomitant impact on the prevalence of HGV in transfusion recipients.
MATERIALS AND METHODS
Patients.The study was performed at a single adult bone marrow transplant center that serves the West Midlands Region of the United Kingdom. Patients who had either been treated for acute leukemia or who had undergone a bone marrow transplant (BMT) procedure were studied. All available patients who were treated and transfused before September 1991 (the date that donor screening for HCV was fully implemented in the United Kingdom) were included in the study. Patients who were treated after this date were recruited consecutively from outpatient clinics. There were no criteria for exclusion from the study. The medical and blood bank records of all patients were reviewed to establish the number of exposures to donor blood components. A donor exposure was defined as a single transfused unit of red blood cells, platelets, fresh frozen plasma, cryoprecipitate, or a platelet apheresis pack. Patient records were also examined to obtain the results of all previous serum aspartate aminotransferase (AST) levels taken during and after treatment. The unit policy is to measure AST levels three times per week during inpatient stays and at every outpatient attendance. For the purposes of this study, the AST level was considered consistently elevated when greater than 35 U/L on two samples at least 6 months apart (upper end of normal range, 30 U/L). At routine follow-up, serum samples were obtained from all patients and assayed for AST. After collection, serum samples were frozen and stored at −20°C before subsequent HGV and HCV testing. Patients gave consent for samples to be taken for research purposes and, before HGV testing but after data collection, all samples were anonymized.
Assays.HGV RNA was extracted from 100 μL of patient serum with Purescript (Gentra Systems Inc, Minneapolis, MN) according to the manufacturer's instructions. cDNA synthesis of 10 μL RNA was performed using random hexamers (Pharmacia Biotech Inc, Piscataway, NJ) and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). PCR was performed using non-nested primers derived from the NS5 region of the HGV genome (77F and 211R; Table 1) and nested primers derived from the NS3 region (S1, Q1, S2, Q2; Table 1).3 Each PCR reaction was performed in a 50 μL volume using Taq DNA polymerase (Promega) for 45 cycles consisting of denaturation for 60 seconds at 95°C, annealing for 75 seconds at 55°C, and extension for 60 seconds at 72°C. PCR products were analyzed on a 2% agarose gel. The expected size of the products from the non-nested PCR were 152 bp and 194 bp for the second round of the nested PCR. Samples that gave positive results in both PCR reactions were considered HGV-PCR positive. No specimens gave indeterminate results, ie, positive in one PCR reaction only. Assays were validated by the inclusion of known positive and negative control samples. Precautions taken to avoid DNA contamination were those described by Kwok et al.8 Sequencing of the nested PCR product in four randomly selected samples confirmed the correct viral target (data not shown).
HGV Primer Sequences
| . | . | Sequence 5′-3′ . |
|---|---|---|
| Non-nested primers | 77F | CTCTTTGTGGTAGTAGCCGAGAGAT |
| 211R | CGAATGAGTCAGAGGACGGGGTAT | |
| Nested primers — outer | S1 | GACGTTGGTGAGATCCCCTT |
| Q1 | CGAAGTTTCCTGTGTACCC | |
| Nested primers — inner | S2 | TGGGCATGGTATCCCCCTCGAGC |
| Q2 | TAGAGAGCGCGTCTGTCGCAC |
| . | . | Sequence 5′-3′ . |
|---|---|---|
| Non-nested primers | 77F | CTCTTTGTGGTAGTAGCCGAGAGAT |
| 211R | CGAATGAGTCAGAGGACGGGGTAT | |
| Nested primers — outer | S1 | GACGTTGGTGAGATCCCCTT |
| Q1 | CGAAGTTTCCTGTGTACCC | |
| Nested primers — inner | S2 | TGGGCATGGTATCCCCCTCGAGC |
| Q2 | TAGAGAGCGCGTCTGTCGCAC |
Anti-HCV testing was performed using Ortho HCV 3.0 ELISA (Ortho Diagnostic Systems, Neckargemünd, Germany). Reactive samples were further investigated using a second enzyme immunoassay (EIA; Monolisa; Sanofi Diagnostic Pasteur, Marne la Coquette, France) and recombinant immunoblot assay (RIBA HCV 3.0 SIA; Ortho Diagnostic Systems). All samples were assessed for the presence of HCV RNA by RT-PCR using a commercial HCV PCR assay (Roche Diagnostic Systems Inc, Branchburg, NJ).
Statistics.The Mann Whitney test and χ2 test were performed using Minitab for Windows (State College, PA).
RESULTS
Sixty patients were identified and studied (patient diagnoses are shown in Table 2). The patients were a mean of 5.4 years from diagnosis and 4.1 years from completion of treatment. At the time of HGV testing, 11 patients were still receiving treatment and 4 patients were in remission over 10 years since completion of therapy. A total of 29 patients (48%) were HGV positive by RT-PCR using both non-nested and nested techniques. Two patients (both in the cohort of 39 patients treated before donor HCV screening) were HCV positive by EIA and RT-PCR; neither was HGV positive. The total number of donor exposures for the whole group was 8,917, with a mean number of donor exposures per individual of 149. There was no significant difference between HGV-positive and -negative patients in respect to donor exposures. When the number of transfused red cell and platelet units were analyzed independently, a trend towards an increased number of transfused red cell units in the HGV-positive group compared with the HGV-negative group was observed. There was no such trend with respect to number of platelet units (Table 3). There was no significant difference in either the time from diagnosis or time from completion of treatment between the HGV-positive and HGV-negative groups (Table 3). Of the 11 patients who were still receiving therapy, 4 were HGV positive. There was no significant difference in time from diagnosis between the HGV-positive and -negative patients in this subgroup.
Diagnoses, HGV Status, and Treatment of the 60 Patients Studied
| Diagnosis . | Total . | HGV Positive . | HGV Negative . | ABMT . | AlloBMT . |
|---|---|---|---|---|---|
| ALL | 10 | 4 | 6 | 3 | 1 |
| AML | 31 | 13 | 18 | 6 | 4 |
| CML | 5 | 2 | 3 | 1 | 4 |
| Hodgkins | 6 | 3 | 3 | 6 | — |
| NHL | 6 | 5 | 1 | 6 | — |
| Myeloma | 2 | 2 | 0 | 2 | — |
| Diagnosis . | Total . | HGV Positive . | HGV Negative . | ABMT . | AlloBMT . |
|---|---|---|---|---|---|
| ALL | 10 | 4 | 6 | 3 | 1 |
| AML | 31 | 13 | 18 | 6 | 4 |
| CML | 5 | 2 | 3 | 1 | 4 |
| Hodgkins | 6 | 3 | 3 | 6 | — |
| NHL | 6 | 5 | 1 | 6 | — |
| Myeloma | 2 | 2 | 0 | 2 | — |
Abbreviations: HGV, hepatitis G virus; ABMT, autologous bone marrow transplantation; AlloBMT, allogeneic bone marrow transplantation; ALL, acute lymphoblastic leukemia; AML, acute myelogenous leukemia; CML, chronic myelogenous leukemia; NHL, non-Hodgkin's lymphomas.
HGV Status and Blood Component Exposure, AST Levels, and Times From Diagnosis and Treatment
| . | HGV Positive . | HGV Negative . | P Value . |
|---|---|---|---|
| . | (n = 29) . | (n = 31) . | . |
| Median time from diagnosis — years | 4.43 | 6.12 | .14 |
| (range) | (0.23-11.3) | (0.05-15.2) | |
| Median time from completion of treatment — years | 3.2 | 4.6 | .18 |
| (range) | (0-10.5) | (0-13.6) | |
| Median donor exposures | 130 | 95 | .48 |
| (range) | (20-685) | (17-483) | |
| Median red blood cell units | 37 | 25 | .12 |
| (range) | (3-96) | (4-88) | |
| Median platelet units | 78 | 84 | .79 |
| (range) | (9-610) | (0-412) | |
| Median concurrent AST level — U/L | 27 | 23 | .28 |
| (range) | (15-77) | (12-393) | |
| Median previous peak AST level — U/L | 72 | 82 | .68 |
| (range) | (17-427) | (13-777) |
| . | HGV Positive . | HGV Negative . | P Value . |
|---|---|---|---|
| . | (n = 29) . | (n = 31) . | . |
| Median time from diagnosis — years | 4.43 | 6.12 | .14 |
| (range) | (0.23-11.3) | (0.05-15.2) | |
| Median time from completion of treatment — years | 3.2 | 4.6 | .18 |
| (range) | (0-10.5) | (0-13.6) | |
| Median donor exposures | 130 | 95 | .48 |
| (range) | (20-685) | (17-483) | |
| Median red blood cell units | 37 | 25 | .12 |
| (range) | (3-96) | (4-88) | |
| Median platelet units | 78 | 84 | .79 |
| (range) | (9-610) | (0-412) | |
| Median concurrent AST level — U/L | 27 | 23 | .28 |
| (range) | (15-77) | (12-393) | |
| Median previous peak AST level — U/L | 72 | 82 | .68 |
| (range) | (17-427) | (13-777) |
Abbreviations: HGV, hepatitis G virus; AST, aspartate aminotransferase.
There were 15 patients with AST levels greater than 35 U/L (normal range, 4 to 30 U/L) at the time of HGV testing, and of these, 8 (53%) were HGV positive and 7 were HGV negative. There was no significant difference in HGV positivity rates between those with normal AST levels and those with raised AST levels. In those patients who had complete records of AST levels available, 42 of 52 (81%) had elevated levels on at least one occasion. Although AST levels as high as 777 U/L were recorded, most elevations were transient, and only 14 patients had persistently elevated levels. Of these 14 patients, 7 were HGV positive. There was no significant difference in peak AST levels between HGV-positive and -negative patients (Table 3). Data on previous AST levels were not available for the remaining 8 patients, 5 of whom were HGV positive.
Analysis of patients by treatment type showed that 20 of 33 (61%) patients who received a BMT were HGV positive compared with 9 of 27 (33%) who had been treated with conventional combination chemotherapy (χ2 = 3.76, P = .036). There was no significant difference between these two treatment groups with respect to donor exposures, time from diagnosis, time from end of treatment, or changes in AST level (Table 4).
Treatment Modalities and Times From Diagnosis and Treatment, Blood Components Transfused, and Concurrent AST Levels
| . | BMT . | No BMT . | P Value . |
|---|---|---|---|
| . | (n = 33) . | (n = 27) . | . |
| HGV positive | 20 | 9 | .036 |
| Median time from diagnosis — years | 5.48 | 4.71 | .71 |
| (range) | (1.19-11.4) | (0.05-15.2) | |
| Median time from completion of treatment — years | 3.86 | 3.88 | .71 |
| (range) | (0-10.5) | (0-13.6) | |
| Median donor exposures | 95 | 116 | .65 |
| (range) | (12-685) | (17-443) | |
| Median red blood cell units | 27 | 30 | .16 |
| (range) | (3-93) | (4-96) | |
| Median platelet units | 84 | 67 | .81 |
| (range) | (9-610) | (0-315) | |
| Median concurrent AST level | 28 | 23 | .10 |
| (range) | (15-393) | (12-94) |
| . | BMT . | No BMT . | P Value . |
|---|---|---|---|
| . | (n = 33) . | (n = 27) . | . |
| HGV positive | 20 | 9 | .036 |
| Median time from diagnosis — years | 5.48 | 4.71 | .71 |
| (range) | (1.19-11.4) | (0.05-15.2) | |
| Median time from completion of treatment — years | 3.86 | 3.88 | .71 |
| (range) | (0-10.5) | (0-13.6) | |
| Median donor exposures | 95 | 116 | .65 |
| (range) | (12-685) | (17-443) | |
| Median red blood cell units | 27 | 30 | .16 |
| (range) | (3-93) | (4-96) | |
| Median platelet units | 84 | 67 | .81 |
| (range) | (9-610) | (0-315) | |
| Median concurrent AST level | 28 | 23 | .10 |
| (range) | (15-393) | (12-94) |
Abbreviations: AST, aspartate aminotransferase; BMT, bone marrow transplantation; HGV, hepatitis G virus.
The data were assessed to see if there was a difference in prevalence of HGV between the cohort of patients who were treated and transfused before the introduction of donor screening for HCV and the cohort who only received transfusions after this date. There was no significant difference in HGV positivity between these two subgroups: 18 of 39 (46%) patients treated before HCV screening were HGV positive compared with 11 of 21 (52%) treated after screening.
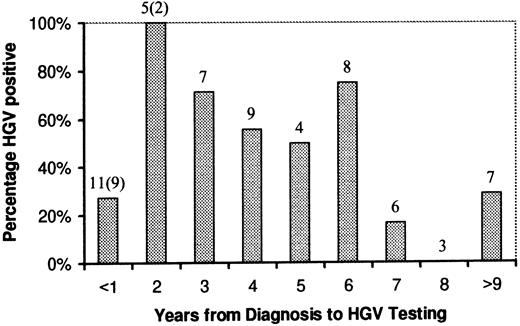
Although there was an apparent trend towards a lower HGV positivity rate with increasing time from diagnosis in those patients who had completed treatment, this was not statistically significant. The percentage of HGV-positive patients with respect to time from diagnosis is shown in Fig 1.
HGV rate related to time from diagnosis. Numbers above columns refer to the number of patients in each group. The numbers in parentheses are patients still undergoing treatment at the time of testing.
HGV rate related to time from diagnosis. Numbers above columns refer to the number of patients in each group. The numbers in parentheses are patients still undergoing treatment at the time of testing.
No patients were on specific antiviral therapy at the time of HGV testing. During the course of treatment, all five of the patients with chronic myeloid leukemia had received therapy with α interferon, but this had been discontinued before BMT and exposure to blood components.
DISCUSSION
The finding that almost half of this multitransfused group of patients are positive by RT-PCR for HGV is further evidence that this virus is transmitted by blood transfusion. Linnen et al3 indicated that the incidence of HGV in volunteer blood donors in the United States is around 1.6%. The prevalence in blood donors in the United Kingdom has recently been reported to be 3.2%.9 If this is a true reflection of the prevalence in the West Midlands donor pool, we would expect that 285 of the 8,917 donor units transfused would be HGV positive. It follows that each of our patients would have been exposed to an average of four HGV-infected units.
The finding of two HCV-positive patients in the cohort treated before the introduction of donor HCV screening is in keeping with previously published results for the UK population.10,11 Although it has been suggested that groups at risk of acquiring HCV are also at risk of HGV,12 screening of blood donors for anti-HCV appeared to have no impact on the prevalence of HGV in our study. This, together with the relatively high donor HGV prevalence, suggests a discordance between HGV and HCV within our population. The mode or modes of HGV transmission to such a relatively high percentage of blood donors is presently unknown. The UK volunteer blood donor population are highly selected, are well motivated, and are well educated regarding self-deferral if they have risk factors (intravenous drug use, etc) for blood-borne infections. Additionally, all donations are screened for the parenterally transmitted viruses, human immunodeficiency virus 1 and 2, hepatitis B, and HCV. This raises the possibility that a nonparenteral route for HGV transmission may be involved.
Retrospective studies suggest that HGV RNA could persist for at least 4 years after infection.3 13 Our data show that HGV may persist in patients for considerably longer; two patients were more than 9 years from the last possible transfusion exposure yet remained HGV-RNA positive.
The patients in this study differ markedly from the UK hemophilia population studied by others, in which the reported prevalence of anti-HCV was 83%, almost all having detectable HCV RNA, but in which only 14% are HGV positive.9 It is possible that the high rate of HGV positivity in our study may be related to immunosuppression at the time of exposure. This might increase the risk of infection and persistence after exposure to HGV-positive blood products. The higher rate of HGV positivity in the more severely immunocompromised BMT subgroup, despite similar levels of transfusion as non-BMT patients, supports this hypothesis. Some patients who are HGV negative may have been infected and may have subsequently cleared the virus, as has been shown in 2 of 12 patients with transfusion-associated hepatitis (non–A-E).3 When reliable assays for HGV antibodies become available, it will be possible to better establish the incidence of infection.
Another possible explanation for the difference in positivity rates between hemophiliacs and our group of patients is that there is a higher viral load in cellular blood components compared with plasma products. Indeed, it has been suggested that HGV is lymphotrophic, because it has been isolated from the mononuclear component of whole blood after the supernatant had been rendered PCR negative by repeated washing.14
The clinical significance of HGV infection in this study is uncertain. The lack of an association between hepatic dysfunction (as measured by serum AST levels) and HGV positivity in this study suggests that it may be of little clinical significance in at least a proportion of those infected. It is recognized that there are several reasons, in addition to viral hepatitis, that may cause raised AST levels in this patient group, such as drug toxicity, veno-occlusive disease, iron overload, and graft-versus-host disease. Although HGV has been implicated in cases of fulminant hepatic failure (FHF ),6 others have noted that it often causes little or no hepatic dysfunction,12,13 and Sallie et al15 found no evidence of HGV in 20 patients undergoing liver transplantation for FHF. There remains the possibility that patients who are HGV positive may have advanced liver disease in the absence of increased serum AST levels because this phenomenon is well recognized in the case of HCV infection.16 A clearer understanding of the clinical significance of this virus may be obtained when the rate of HGV positivity is established in patients with cryptogenic cirrhosis and hepatocellular carcinoma, because these conditions represent the end result of significant chronic liver disease. Given the high rate of HGV positivity in multitransfused hematology patients, the possibility that it may cause chronic liver disease as is evident for HCV is clearly of great concern, and the need for further studies to establish the natural history of this virus is urgent.
ACKNOWLEDGMENT
We thank Drs P. Karayiannis, J. Pickering, and Prof H. Thomas (St Mary's Hospital Medical School, London, UK) for advice and for undertaking nucleic acid sequencing.
Address reprint requests to Susan J. Skidmore, PhD, Public Health Laboratory (Regional Virus Laboratory), Birmingham Heartlands Hospital, Birmingham B9 5SS, UK.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal