Abstract
The cell surface zinc metalloproteinase CD10/neutral endopeptidase 24.11 ([NEP] neprilysin) functions as part of a regulatory loop to control local concentrations of peptide substrates and associated peptide-mediated signal transduction. The physiologic role of the enzyme depends on available substrates in specific organs and cell types. Although CD10/NEP is expressed on a restricted subset of normal and malignant lymphoid progenitors, the enzyme is also expressed by a variety of epithelial cells. To explore the mechanism of tissue-specific expression of this regulatory enzyme, we characterized the major (type 2) CD10/NEP promoter and identified three functionally active transcription factor binding sites (regions I to III). CBF/NF-Y binds to the inverted CCAAT box in region I, whereas a second positive and a third negative factor bind to regions II and III, respectively. Although region I is required for maximal CD10/NEP-driven luciferase activity in the examined epithelial cell lines, this region is not required for maximal activity in the evaluated lymphoid cell lines. The apparent tissue-specific differences in requirements for region I (and CBF/NF-Y) are of particular interest because lymphoid and epithelial cells express alternatively spliced versions of CBF/NF-Y that differ in biologic activity.
THE CD10/neutral endopeptidase 24.11 ([NEP] EC 3.4.24.11, neprilysin) is a cell surface zinc metalloproteinase that regulates the biologic activity of peptide substrates by reducing the local concentrations available for receptor binding and signal transduction.1 CD10/NEP is a member of a newly identified gene family that includes the endothelin-converting enzyme and the Kell blood group protein.2,3 These proteins have striking sequence similarity within the C-terminal third of their putative extracellular domains, although their N-terminal regions are less closely related.2 3
CD10/NEP is expressed in a highly restricted fashion on normal and malignant lymphoid progenitors.4-6 However, the enzyme is also expressed on terminally differentiated granulocytes and a variety of nonhematopoietic cells, including bronchial epithelial cells, bone marrow stromal elements, renal proximal tubular cells, endometrial cells, breast myoepithelium, fetal intestine, and certain solid-tumor cell lines.1
Because CD10/NEP cleaves small peptides on the amino-terminal side of hydrophobic amino acids, there are many known CD10/NEP substrates including substance P, met- and leu-enkephalin, FMLP, the bombesin-like peptides, atrial natriuretic factor, endothelin, and oxytocin.7-14 The physiologic role of the enzyme depends on available substrates in specific organs and cell types. For example, CD10/NEP reduces enkephalin-mediated analgesia,15 atrial natriuretic factor–mediated hypertension and diuresis,10 endothelin-regulated menstrual bleeding,14,16 and peptide-mediated inflammatory responses.8 17-20
The enzyme also regulates peptide-mediated cellular proliferation. CD10/NEP hydrolyzes bombesin-like peptides that are potent mitogens for fibroblasts and normal bronchial epithelial cells and essential autocrine growth factors for many small-cell carcinomas of the lung.21 CD10/NEP also regulates the growth and maturation of early lymphoid progenitors.22,23 Although the CD10/NEP peptide substrate for lymphoid progenitors has not yet been identified, the enzyme clearly modulates stromal cell–dependent early B-lymphopoiesis.22 23
Although CD10/NEP functions to regulate local concentrations of peptide substrates, the enzyme itself is regulated by a variety of stimuli. For example, phorbol ester treatment of acute lymphoblastic leukemia cells, epithelial cells, and granulocytes reduces CD10/NEP protein and transcripts,24-26 whereas treatment with dexamethasone, progesterone, and additional factors such as FMLP, granulocyte-macrophage colony-stimulating factor, tumor necrosis factor, or C5a increase the enzyme levels.16,20,27 28
The important regulatory roles of CD10/NEP and its known relationship to cellular activation and proliferation prompted us to characterize its 5′ sequence. In our previous studies, the CD10/NEP gene was found to include two different 5′ (noncoding) exons (exons 1 and 2A/B) that splice into a common coding exon (originally identified as exon 3).29,30 Three types of CD10/NEP transcripts result from alternative splicing of these specific 5′ untranslated regions. In type 1 transcripts, exon 1 splices directly into the common coding exon.29 In types 2A and 2B transcripts, exon 2 splices into the common coding exon using either an internal or 3′ exon 2 splice site.29 Two additional noncoding exons have been described.30,31 These noncoding exons, which are located proximal30 and distal31 to exon 2, also splice into the common coding exon.30 31
In previous studies, type 2 transcripts were more abundant in the majority of human CD10/NEP-positive (CD10/NEP+) cell lines and organs.32 The abundance of type 1 transcripts was more variable, with the highest type 1 levels in fetal thymus and certain lymphoblastic leukemia and glioblastoma cell lines.32 In similar analyses of CD10/NEP transcripts in the rat, type 2 transcripts were more abundant in many tissues, although type 1 transcripts predominated in specific regions of the brain and spinal cord.33
CD10/NEP type 1 and 2 transcripts are controlled by separate regulatory elements that are both characterized by the presence of multiple transcription initiation sites and the absence of classic TATA boxes and consensus initiation elements.32 The GC-rich type 2 regulatory region also contains an inverted CCAAT box, a motif that has been implicated in the transcription initiation of cell cycle control genes.32 Because the type 2 promoter is more active in the majority of human lymphoid and epithelial cell lines examined to date, we further characterized this regulatory element and identified three functionally relevant transcription factor binding sites, one of which is CBF/NF-Y.
MATERIALS AND METHODS
Cell lines.Cell lines used to assess CD10/NEP-driven luciferase activity included the SV40-transformed human fetal bronchial epithelial cell line FHTE56, the human acute lymphoblastic leukemia cell line Nalm-6, the human Burkitt's lymphoma cell line Raji (ATCC CCL86), and the human non–small-cell lung cancer cell line Calu-1 (ATCC HTB54).34 FHTE56 was maintained on fibronectin (1 mg/100 mL; GIBCO BRL, Gaithersburg, MD)/collagen (1% vitrogen; Celtrix Laboratory, Palo Alto, CA)–coated plates in Dulbecco's modified Eagle's medium (DMEM)/10% fetal calf serum (FCS).34 Nalm-6 and Raji were grown in RPMI/10% FCS, and Calu-1 was cultured in DMEM/10% FCS.
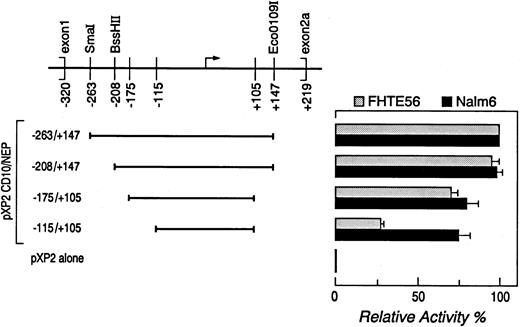
Determination of CD10/NEP promoter activity by luciferase activity.A series of CD10/NEP genomic fragments containing overlapping segments of 5′ CD10/NEP sequence were subcloned into the promoterless luciferase vector pXP2.32 CD10/NEP genomic fragments were obtained from previously characterized larger CD10/NEP-pXP2 constructs32 by restriction endonuclease digestion (fragment −263/+147, SmaI/EcoO109I; fragment −208/+147, BssHII/EcoO1091) or polymerase chain reaction (PCR) with sequence-specific oligonucleotide primers (fragment −175/+105, sense primer 5′-gcttggctgggctctca-3′ and antisense primer 5′-agggtcctgggcgctcg-3′; fragment −115/+105, sense primer 5′-gtgtcggctcagcagcc-3′ and antisense primer 5′-agggtcctgggcgctcg-3′; Fig 1). The resulting PCR fragments were sequenced to rule out Taq polymerase–induced mutations in CD10/NEP sequence.
Deletion analysis of the type 2 CD10/NEP promoter. (Left) CD10/NEPpXP2 deletion constructs. Deletion constructs containing progressively shorter segments of the type 2 CD10/NEP promoter in the promoterless luciferase vector pXP2 are shown. Positions of the indicated constructs are relative to the major initiation of transcription site (). (Right) Luciferase assays of CD10/NEPpXP2 deletion constructs. The resulting CD10/NEPpXP2 constructs and CMV-GH internal controls were cotransfected into Nalm-6 cells and FHTE56 cells to analyze CD10/NEP-driven luciferase activity. In all assays, luciferase activity was normalized for transfection efficiency by evaluating supernatants from the transient transfections for simultaneous GH secretion (ng GH secretion/mL supernatant). Promoter activity (RLUs) and % maximum CD10/NEP-driven activity represent the mean ± SE for two separate experiments performed in triplicate: FHTE56 v, Nalm-: −263/+147, 742 (100%) v 639 (100%); −208/+147, 709(95% ± 5%) v 621(98% ± 3%); −175/+105, 538(71% ± 3%) v 554(80% ± 7%); −1151/+105, 210(27% ± 2%) v 519(75% ± 7%); P < .001. pXP2 alone, 66 v 35, respectively. The differences in CD10/NEP-driven luciferase activity of CD10/NEP(−115/+105)pXP2 in FHTE56 and Nalm-6 cells were analyzed using a one-sided Student's t-test.
Deletion analysis of the type 2 CD10/NEP promoter. (Left) CD10/NEPpXP2 deletion constructs. Deletion constructs containing progressively shorter segments of the type 2 CD10/NEP promoter in the promoterless luciferase vector pXP2 are shown. Positions of the indicated constructs are relative to the major initiation of transcription site (). (Right) Luciferase assays of CD10/NEPpXP2 deletion constructs. The resulting CD10/NEPpXP2 constructs and CMV-GH internal controls were cotransfected into Nalm-6 cells and FHTE56 cells to analyze CD10/NEP-driven luciferase activity. In all assays, luciferase activity was normalized for transfection efficiency by evaluating supernatants from the transient transfections for simultaneous GH secretion (ng GH secretion/mL supernatant). Promoter activity (RLUs) and % maximum CD10/NEP-driven activity represent the mean ± SE for two separate experiments performed in triplicate: FHTE56 v, Nalm-: −263/+147, 742 (100%) v 639 (100%); −208/+147, 709(95% ± 5%) v 621(98% ± 3%); −175/+105, 538(71% ± 3%) v 554(80% ± 7%); −1151/+105, 210(27% ± 2%) v 519(75% ± 7%); P < .001. pXP2 alone, 66 v 35, respectively. The differences in CD10/NEP-driven luciferase activity of CD10/NEP(−115/+105)pXP2 in FHTE56 and Nalm-6 cells were analyzed using a one-sided Student's t-test.
Transient transfections were performed as previously described.32 35 In brief, 5 × 107 Nalm-6 or Raji or 2 × 107 FHTE56 or Calu-1 cells were electroporated with 20 μg pXP2, cytomegalovirus (CMV)-pXP2, or CD10/NEPpXP2 and 2 μg CMV-human growth hormone (GH) in Iscove's modified Dulbecco's medium or DMEM at 300V or 250V and 960 μF. Luciferase activity was measured in relative light units (RLUs) 8 hours posttransfection using a luminometer (Analytical Luminescence Laboratory, San Diego, CA), and GH level was measured by radioimmunoassay (Nichols Institute Diagnostics, San Juan Capistrano, CA). Thereafter, RLUs from individual transfections were normalized for transfection efficiency by standardizing RLUs for GH production driven by the CMV-GH internal control.
DNase I footprinting.Nuclear extracts for DNase I footprinting were prepared as previously described.36 In brief, nuclei were isolated by suspending cells in buffer A (10 mmol/L HEPES, pH 7.8, 1.5 mmol/L MgCl2 , 10 mmol/L KCl, 0.5 mmol/L DTT, and 0.5 mmol/L PMSF) and applying 10 strokes in a Dounce homogenizer using pestle A. The isolated nuclei were then washed and resuspended in buffer B (10 mmol/L HEPES, pH 7.8, 2 mmol/L MgCl2 , 50 mmol/L KCl, 0.1 mmol/L EDTA, 10% glycerol, 0.15 mmol/L spermine, 0.5 mmol/L spermidine, 1 mmol/L DTT and 0.4 mmol/L PMSF). Thereafter, the nuclear protein was extracted with vol 4-mol/L (NH4)2SO4 and centrifuged at 30,000 rpm for 60 minutes. The protein in the supernatant was precipitated by 0.3 g/mL (NH4)2SO4 , pelleted, resuspended in buffer C (20 mmol/L HEPES, pH 7.8, 100 mmol/L KCl, 0.2 mmol/L EDTA, 10% glycerol, 0.5 mmol/L DTT, and 0.5 mmol/L PMSF), and dialyzed against buffer C for 4 hours.
A 314-bp fragment extending from bp −208 to +107 of the type 2 CD10/NEP promoter was obtained by digesting CD10/NEP-263/+147pXP2 with BssHII and EagI (Fig 1). Thereafter, the CD10/NEP bp −208/+107 fragment was 32P-labeled as described previously.37
DNA-binding reactions were performed in a 20-μL volume with 1 × 105 cpm labeled DNA fragment, 4 μg ds-poly(dI-dC) (Pharmacia Biotech, Piscataway, NJ), and 100 μg nuclear protein in a final buffer concentration of 10 mmol/L HEPES, pH 8.0, 30 mmol/L KCl, 5 mmol/L MgCl2 , 1 mmol/L EDTA, 12% glycerol, 0.5 mmol/L DTT, and 0.2 mmol/L PMSF. Reactions were incubated for 60 minutes on ice followed by 60 seconds' digestion at room temperature with a series of concentrations of freshly diluted DNase I (1 to 50 μg/mL); the optimal concentration of DNase I was 6 μL of 10 μg/mL (Worthington Biochemical, Freehold, NJ). DNase I digestion reactions were stopped by 100 μL stop mixture (20 mmol/L Tris, pH 7.5, 20 mmol/L EDTA, 0.5% sodium dodecyl sulfate (SDS), and 100 μg/mL proteinase K). Thereafter, the samples were incubated at 37°C for 30 minutes, extracted with phenol/chloroform/isoamyl alcohol, precipitated with ethanol, and analyzed on a 6% polyacrylamide sequencing gel.
Electrophoretic mobility shift assay.Electrophoretic mobility shift assay (EMSA) was performed as previously described.38 Single-stranded complementary sense and antisense oligonucleotides from regions I, II, and III (region I, sense 5′ accaggccccgtgcgctcattggtcgggat 3′; region II, sense 5′ cttctcccgaatcccactggtgagtcccaggagagcgagct 3′ region III, sense 5′ gagggagaaaggtccaaagggcgcgacgccc 3′ ). CBF/NF-Y (sense 5′-cgtctccaccaatgggagggctgggc-3′ ),39 C/EBP (sense 5′-tgggaagattgagcaatctaagag-3′ ),40 and CTF/NF1 (sense 5′-cctttggcatgctgccaatatg-3′ )41 were synthesized and used to generate double-stranded oligonucleotide probes. In brief, complementary sense and antisense single-stranded oligonucleotide probes were heated together at 95°C for 5 minutes and slowly cooled to room temperature. Thereafter, double-stranded oligonucleotides were 32P-labeled using polynucleotide kinase.
DNA-binding reactions were performed with 1 × 104 cpm probe, 2 μg ds-poly(dI-dC), and 5 μg nuclear protein in 10 mmol/L HEPES, pH 8.0, 30 mmol/L KCl, 5 mmol/L MgCl2 , 1 mmol/L EDTA, 12% glycerol, 0.5 mmol/L DTT, and 0.2 mmol/L PMSF. Reactions were incubated on ice for 15 minutes and subsequently electrophoresed on a 5% polyacrylamide gel in 0.5 × TBE at 4°C. In selected experiments, 100-fold molar excess unlabeled double-stranded oligonucleotides from CD10/NEP regions I, II, and III or CBF-NF-Y, C/EBP, or CTF/NF1 were also added to the binding reactions. In additional supershift experiments, 1 μL anti-CBF-A antibody42 or anti-Oct-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) was added to the binding reaction mixture 15 minutes before addition of the probe.
Site-directed mutagenesis.Mutated type 2 CD10/NEP promoter fragments were generated by PCR38 using CD10/NEP(−263/+147)pBluescript (Stratagene, La Jolla, CA) as template. The antisense M13 forward primer (5′-cgacgttgtaaaacgacggccagtg-3′ ) and regions I, II, or III sense primers (5′-ctagatctagtggtcgggatgtgtc-3′, I-F; 5′-ctagatctagcccaggagagcgagc-3′, II-F; or 5′-ctagatctagaaagggcgcgacgcc-3′, III-F) were used to generate PCR fragments I-F, II-F, or III-F. The sense M13 reverse primer 5′-ggaaacagctatgaccatgattacg-3′ and regions I, II, or III antisense primers (5′-ctagatctagggggccgggtggcag-3′, I-R; 5′-ctagatctaggggattcgggagaag-3′, II-R; or 5′-ctagatctagccctcagctcgctct-3′, III-R) were used to generate PCR fragments I-R, II-R, or III-R. PCR fragments I-F, II-F, and III-F were subsequently digested with BgIII and KpnI, and PCR fragments I-R, II-R, and III-R were digested with BamHI and BgIII. Thereafter, digested PCR fragments I-F and I-R, II-F and II-R, and III-F and III-R were ligated with BamHI/KpnI-digested pXP2 vector to generate the mutated constructs I-M, II-M, or III-M. The sequences of mutated constructs were confirmed before their use in transient transfection assays.
Immunodetection of CBF-B/NF-YA isoforms by Western blot.Nuclear extracts (50 μg) from Nalm-6, Raji, FHTE56, and Calu 1 cells were size-fractionated on 10% SDS–polyacrylamide gels and transferred to Immobilon-P membranes (Millipore Corp, Bedford, MA). The membranes were initially incubated in blocking buffer (10 mmol/L Tris, pH 7.5, 150 mmol/L NaCl, 0.01% Tween 20%, and 5% dry milk) for 1 hour and incubated thereafter with an affinity-purified antiserum directed against the C-terminal peptide of CBF-B/NF-YA43 (gift from R. Mantovani, University of Milan, Milan, Italy). After extensive washes, the membranes were incubated with peroxidase-conjugated anti-rabbit IgG (Amersham) and developed using enhanced chemiluminescence (Amersham).
Transactivation assay.Transactivation studies were performed by cotransfecting Nalm-6 or FHTE56 cells with 20 μg wild-type CD10/NEPpXP2 (−263/+147) or mutant I-M (−263/+147) and 40 μg of either the short or long form of NF-YA in the pSAS eukaryotic expression vector44 (Montavani R., unpublished data) and 1 μg CMV-GH. Luciferase activity was measured 48 hours posttransfection and normalized for GH production.
RESULTS
Specific Type 2 CD10/NEP Promoter Sequences Are Required for Activity in FHTE56 and Nalm-6 Cells
Deletion analysis.In previous studies, the smallest CD10/NEP type 2 fragment with optimal promoter activity included bp −263/+14732 (Fig 1). This minimal type 2 promoter was active in CD10/NEP+ cells, including the bronchial epithelial cell line FHTE56 and the acute lymphoblastic leukemia cell line Nalm-6,32 and inactive in CD10/NEP− cells such as the EBV-transformed lymphoblastoid cell line, Laz 388 (data not shown).
To further delineate the sequences within the minimal type 2 promoter that are required for its activity, we generated a series of deletion constructs containing progressively shorter segments of the type 2 CD10/NEP promoter in a promoterless luciferase vector (pXP2). The resulting CD10/NEPpXP2 constructs and CMV-GH internal controls were cotransfected into Nalm-6 cells and FHTE56 cells to analyze CD10/NEP-driven luciferase activity in representative CD10/NEP+ lymphoid and epithelial cell types (Fig 1). In all assays, luciferase activity was normalized for transfection efficiency by evaluating supernatants from the transient transfections for simultaneous GH secretion.
A CD10/NEPpXP2 construct containing bp −263 to +147 had maximal activity in both FHTE56 cells and Nalm-6 cells (100% baseline value). Stepwise deletions of bp −263 to −175 only reduced CD10/NEP-driven luciferase activity to 71% ± 3% and 80% ± 7% of baseline values in FHTE56 and Nalm-6 cells (Fig 1). In contrast, further deletion of bp −175 to −115 reduced CD10/NEP-driven luciferase activity to 27% ± 2% of baseline values in FHTE56 cells. However, the same deletion (bp −175 to −115) did not significantly reduce CD10/NEP-driven luciferase activity in Nalm-6 cells (75% ± 7% baseline values) (Fig 1). The differences between the effects of deleting bp −175 to −115 in FHTE56 and Nalm-6 cells were highly significant (P < .001), suggesting that regulatory elements within bp −175 to −115 play different roles in the two cell types.
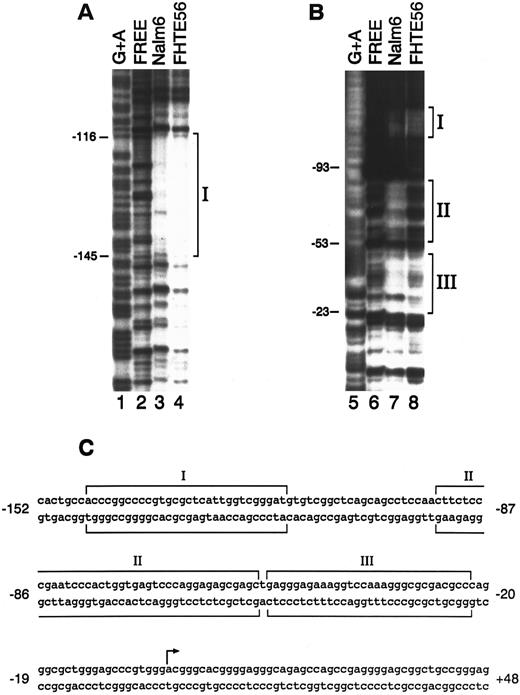
DNase I footprinting.To further define the cis-acting elements and trans-acting factors that control CD10/NEP type 2 promoter activity in epithelial and lymphoid cells, we performed DNase I footprinting with a DNA fragment extending from bp −208 to +107 (Fig 1) and nuclear proteins from FHTE56 and Nalm-6 cells. Three regions of the type 2 CD10/NEP promoter interacted with nuclear proteins from FHTE56 and Nalm-6 cells: region I, bp −145/−116; region II, bp −93/−53; and region III, bp −53/−23. Of note, region I (bp −145/−116) is located within the CD10/NEP promoter sequence required for maximal activity in FHTE56 cells; in contrast, regions II and III are 3′ to this area (Figs 1 and 2). Whereas regions I and III interacted with nuclear proteins from both Nalm-6 and FHTE56 cells, region II was only clearly protected by nuclear proteins from Nalm-6 cells (Fig 2).
DNase I footprinting analysis of the type 2 CD10/NEP promoter. The 314-bp fragment extending from bp −208 to +107 of the CD10/NEP promoter was 32P-labeled on the noncoding strand (A) or coding strand (B) and incubated with no nuclear protein (free, lanes 2 and 6), 100 μg Nalm-6 nuclear protein (lanes 3 and 7), or FHTE56 nuclear protein (lanes 4 and 8). Molecular weight markers (lanes 1 and 5) were generated by G+A-specific Maxam-Gilbert cleavage. Numbers at the left of each panel indicate nucleotides upstream(−) or downstream(+) of the major transcription initiation site (+1). Sequences protected from DNase I digestion are indicated by brackets. (C) Sequence of the CD10/NEP promoter and three protected regions. The arrow delineates the major transcription initiation site (+1).24
DNase I footprinting analysis of the type 2 CD10/NEP promoter. The 314-bp fragment extending from bp −208 to +107 of the CD10/NEP promoter was 32P-labeled on the noncoding strand (A) or coding strand (B) and incubated with no nuclear protein (free, lanes 2 and 6), 100 μg Nalm-6 nuclear protein (lanes 3 and 7), or FHTE56 nuclear protein (lanes 4 and 8). Molecular weight markers (lanes 1 and 5) were generated by G+A-specific Maxam-Gilbert cleavage. Numbers at the left of each panel indicate nucleotides upstream(−) or downstream(+) of the major transcription initiation site (+1). Sequences protected from DNase I digestion are indicated by brackets. (C) Sequence of the CD10/NEP promoter and three protected regions. The arrow delineates the major transcription initiation site (+1).24
Nuclear Proteins From FHTE56 and Nalm-6 Cells Interact With Regions I, II, and III of the CD10/NEP Promoter
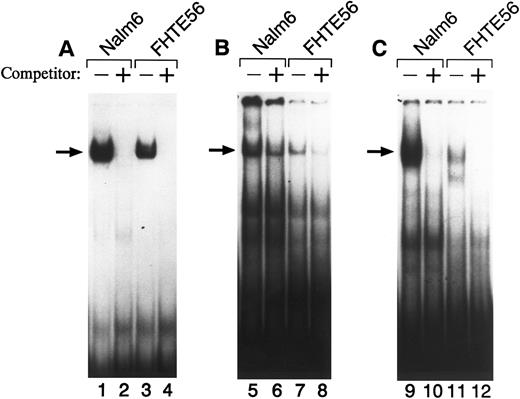
To further characterize factor(s) binding to regions I, II, and III in FHTE56 and Nalm-6 cells, we performed EMSAs (Fig 3). When a 32P-labeled double-stranded region I oligonucleotide (bp −145 through −116) was used as a probe, a major shifted band was detected with nuclear proteins from both Nalm-6 and FHTE56 cells (Fig 3A). Excess unlabeled region I probe eliminated the binding of radiolabeled region I probe to the major shifted band, confirming the specificity of the interaction between region I and the Nalm-6 and FHTE56 nuclear proteins (Fig 3A).
EMSA of the type 2 CD10/NEP promoter with wild-type region I (A), II (B), or III (C) probes. Double-stranded oligonucleotides from regions I, II, or III were 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 (lanes 1, 2, 5, 6, 9, and 10) or FHTE56 (lanes 3, 4, 7, 8, 11, and 12) in the absence (−) or presence (+) of 100-fold molar excess unlabeled self-oligonucleotide. Arrows identify the major shifted bands.
EMSA of the type 2 CD10/NEP promoter with wild-type region I (A), II (B), or III (C) probes. Double-stranded oligonucleotides from regions I, II, or III were 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 (lanes 1, 2, 5, 6, 9, and 10) or FHTE56 (lanes 3, 4, 7, 8, 11, and 12) in the absence (−) or presence (+) of 100-fold molar excess unlabeled self-oligonucleotide. Arrows identify the major shifted bands.
A 32P-labeled double-stranded region II oligonucleotide probe (bp −93 through −53) also bound to a nuclear protein from Nalm-6 cells. Of note, the region II oligonucleotide probe also bound to a similar-sized nuclear protein from FHTE56 cells (Fig 3B), although region II was not clearly protected in the less sensitive FHTE56 DNase I footprinting assays (Fig 2B). A 32P-labeled double-stranded region III oligonucleotide probe also bound to a nuclear protein in Nalm-6 and FHTE56 cells and an additional smaller nuclear protein in FHTE56 cells (Fig 3C).
Mutations in Regions I, II, and III Abolish the Binding of Factors to the CD10/NEP Type 2 Promoter
To further characterize the functional significance of the identified binding sites in the CD10/NEP promoter, region I, II, and III binding sites were each mutated (Fig 4A). Thereafter, the affinity of the mutated binding sites for Nalm-6 nuclear proteins was analyzed and compared with that of wild-type binding sites in EMSAs (Fig 4B to D); the affinity of the mutated binding sites for FHTE56 nuclear proteins was similarly analyzed (data not shown). Whereas wild-type 32P-labeled region I, II, and III probes bound the indicated Nalm-6 nuclear proteins, mutated region I, II, and III probes did not bind Nalm-6 nuclear proteins (Fig 4B to D). Furthermore, unlabeled mutated region I, II, or III oligonucleotide probes did not effectively compete with 32P-labeled wild-type probes under conditions in which unlabeled wild-type probes inhibited the binding of 32P-labeled probes to Nalm-6 nuclear proteins (Fig 4B to D).
EMSA of the type 2 CD10/NEP promoter with mutated region I, II, or III probes. (A) Sequences of wild-type and mutant region I, II, or III oligonucleotide probes. The 10-bp mutations in region I, II, and III oligonucleotide probes are indicated. (B, C, and D) EMSAs. A double-stranded oligonucleotide of wild-type (W) region I (B), region II (C), or region III (D) was 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 in the absence (−, lanes 1, 5, and 9) or presence of 100-fold molar excess unlabeled W (lanes 2, 6, and 10) or mutated ([M] lanes 3, 7, and 11) oligonucleotide. Double-stranded mutated region I, II, or III oligonucleotide probes (M) were also 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 (lanes 4, 8, and 12). Arrows identify the major shifted bands.
EMSA of the type 2 CD10/NEP promoter with mutated region I, II, or III probes. (A) Sequences of wild-type and mutant region I, II, or III oligonucleotide probes. The 10-bp mutations in region I, II, and III oligonucleotide probes are indicated. (B, C, and D) EMSAs. A double-stranded oligonucleotide of wild-type (W) region I (B), region II (C), or region III (D) was 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 in the absence (−, lanes 1, 5, and 9) or presence of 100-fold molar excess unlabeled W (lanes 2, 6, and 10) or mutated ([M] lanes 3, 7, and 11) oligonucleotide. Double-stranded mutated region I, II, or III oligonucleotide probes (M) were also 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 (lanes 4, 8, and 12). Arrows identify the major shifted bands.
Mutations in Regions I, II, and III Alter CD10/NEP-Driven Luciferase Activity
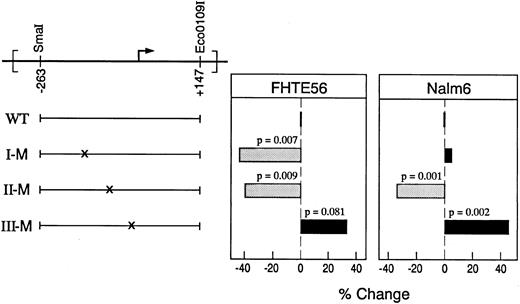
To further assess the roles of the identified binding sites in the type 2 CD10/NEP promoter, the indicated region I, II, and III mutations (Fig 4A) were separately generated in the CD10/NEP(−263/+147)pXP2 construct (Fig 5). The indicated region I mutation was chosen to overlap an identified inverted CCAAT sequence (Fig 4A). The indicated region II and III mutations were placed in the middle of the sequence most clearly protected in Nalm-6 cells (Fig 2B). Thereafter, CD10/NEP-driven luciferase activities of wild-type and mutated constructs were compared in transient transfection assays of FHTE56 and Nalm-6 cells.
Functional analysis of region I, II, and III mutations in the type 2 CD10/NEP promoter. (Left) Mutated CD10/NEP(−263/+147)pXP2 constructs. Wild-type CD10/NEP (−263/+147)pXP2 construct (WT) and mutated region I (I-M), II (II-M), and III (III-M) constructs are shown. (Right) Luciferase assays of mutated CD10/NEP(−263/+147) constructs. The indicated WT and mutated CD10/NEP (−263/+147)pXP2 constructs were cotransfected with CMV-GH plasmid into FHTE56 and Nalm-6 cells. Resulting luciferase activities were measured and normalized for transfection efficiency by standardizing for GH production driven by the CMV-GH internal control. Promoter activity (RLU) represents the mean ± SE for two separate experiments performed in duplicate: FHTE56 v Nalm-6: WT, 1,293 ± 123 (100%) v 245 ± 12 (100%); I-M, 721 ± 69 (56%) v 257 ± 18 (105%); II-M, 774 ± 57 (60%) v 162 ± 8 (66%); III-M, 1,716 ± 160 (133%) v 357 ± 18 (146%). Differences between promoter activity driven by WT and mutated constructs were evaluated using a one-sided Student's t-test.
Functional analysis of region I, II, and III mutations in the type 2 CD10/NEP promoter. (Left) Mutated CD10/NEP(−263/+147)pXP2 constructs. Wild-type CD10/NEP (−263/+147)pXP2 construct (WT) and mutated region I (I-M), II (II-M), and III (III-M) constructs are shown. (Right) Luciferase assays of mutated CD10/NEP(−263/+147) constructs. The indicated WT and mutated CD10/NEP (−263/+147)pXP2 constructs were cotransfected with CMV-GH plasmid into FHTE56 and Nalm-6 cells. Resulting luciferase activities were measured and normalized for transfection efficiency by standardizing for GH production driven by the CMV-GH internal control. Promoter activity (RLU) represents the mean ± SE for two separate experiments performed in duplicate: FHTE56 v Nalm-6: WT, 1,293 ± 123 (100%) v 245 ± 12 (100%); I-M, 721 ± 69 (56%) v 257 ± 18 (105%); II-M, 774 ± 57 (60%) v 162 ± 8 (66%); III-M, 1,716 ± 160 (133%) v 357 ± 18 (146%). Differences between promoter activity driven by WT and mutated constructs were evaluated using a one-sided Student's t-test.
Region I.Mutation of the region I binding site significantly reduced CD10/NEP-driven luciferase activity in FHTE56 cells. However, the region I mutation hads no effect on CD10/NEP-driven luciferase activity in Nalm-6 cells (Fig 5). These data are consistent with the original analyses in which deletion of region I reduced CD10/NEP-driven luciferase activity in FHTE56 epithelial cells but not in Nalm-6 lymphoid cells (Fig 1, CD10/NEP [−115/+105] pXP2).
To further assess the apparent tissue-specific differences in region I activity, the wild-type and mutated region I constructs were analyzed in additional CD10/NEP+ lymphoid (Raji) and epithelial (Calu-1) cell lines. Although the region I mutation had no effect on CD10/NEP-driven luciferase activity in Raji cells (1% reduction, P = NS), this mutation significantly reduced CD10/NEP-driven luciferase activity in Calu-1 cells (58% reduction, P = .007) (data not shown). Taken together, these data suggest that the region I regulatory element is required for CD10/NEP activity in epithelial cell lines such as FHTE56 and Calu-1 but not in B-lymphoid cell lines such as Nalm-6 and Raji.
Regions II and III.Mutation of the region II binding site significantly reduced CD10/NEP-driven luciferase activity in both FHTE56 and Nalm-6 cells. In marked contrast, mutation of the region III binding site increased CD10/NEP-driven luciferase activity in both cell types (Fig 5). These data suggest that region II is a positive regulatory element and region III a negative regulatory element in FHTE56 and Nalm-6 cells (Fig 5).
The Region I Binding Factor is CBF/NF-Y
To identify potential region I binding factors, the region I sequence was analyzed for known transcription factor binding sites. As previously reported,32 region I contains an inverted CCAAT sequence at bp −127 to −123. This inverted CCAAT sequence is interrupted by the 10-bp mutation introduced in region I (Fig 4), suggesting that region I binding factors might interact with the inverted CCAAT sequence.
Mammalian transcription factors that bind to CCAAT elements include C/EBP,40 CTF/NF1,41 and CBF/NF-Y.39,45 46 To determine whether one of these transcription factors binds to region I of the CD10/NEP promoter, we performed additional EMSAs in which 32P-labeled double-stranded region I oligonucleotide probe was incubated with Nalm-6 or FHTE56 nuclear protein in the presence or absence of excess unlabeled CTF/NF1, C/EBP, or CBF/NF-Y double-stranded oligonucleotide probes (Fig 6).
(A) EMSAs of CD10/NEP region I in the presence or absence of oligonucleotides from known CCAAT binding factors. A double-stranded region I oligonucleotide was 32P-labeled and incubated with 5 μg Nalm-6 nuclear protein in the absence (−) (lane 1) or presence of 100-fold molar excess of the indicated unlabeled double-stranded oligonucleotides: CBF/NF-Y, lane 2; C/EBP, lane 3; CTF/NFI, lane 4; self, lane 5. (B) EMSAs of CD10/NEP region I in the presence or absence of a CBF/NF-Y oligonucleotide or antibody. 32P-labeled double-stranded region I oligonucleotide was also incubated with Nalm-6 nuclear protein in the absence (−) (lane 6) or presence of anti–CBF-A [NF-Y] antibody (lane 7) or anti-OCT1 antibody (lane 8), or with FHTE56 nuclear protein in the absence (−) (lane 9) or presence of unlabeled self-oligonucleotide probe (lane 10), unlabeled CBF/NF-Y oligonucleotide probe (lane 11), anti–CBF-A [NF-Y] antibody (lane 12), or anti-Oct1 antibody (lane 13). Upper arrow indicates supershifted bands and lower arrow indicates major shifted bands in the absence of specific antibodies. (C) Electrophoretic mobility of region I/nuclear protein complexes from lymphoid and epithelial cells. Region I/lymphoid nuclear protein complexes (Nalm-6 [lane 14] and Raji [lane 16]) migrate slightly faster than region I/epithelial nuclear protein complexes (FHTE56 [lane 15] and Cal-1 [lane 17]).
(A) EMSAs of CD10/NEP region I in the presence or absence of oligonucleotides from known CCAAT binding factors. A double-stranded region I oligonucleotide was 32P-labeled and incubated with 5 μg Nalm-6 nuclear protein in the absence (−) (lane 1) or presence of 100-fold molar excess of the indicated unlabeled double-stranded oligonucleotides: CBF/NF-Y, lane 2; C/EBP, lane 3; CTF/NFI, lane 4; self, lane 5. (B) EMSAs of CD10/NEP region I in the presence or absence of a CBF/NF-Y oligonucleotide or antibody. 32P-labeled double-stranded region I oligonucleotide was also incubated with Nalm-6 nuclear protein in the absence (−) (lane 6) or presence of anti–CBF-A [NF-Y] antibody (lane 7) or anti-OCT1 antibody (lane 8), or with FHTE56 nuclear protein in the absence (−) (lane 9) or presence of unlabeled self-oligonucleotide probe (lane 10), unlabeled CBF/NF-Y oligonucleotide probe (lane 11), anti–CBF-A [NF-Y] antibody (lane 12), or anti-Oct1 antibody (lane 13). Upper arrow indicates supershifted bands and lower arrow indicates major shifted bands in the absence of specific antibodies. (C) Electrophoretic mobility of region I/nuclear protein complexes from lymphoid and epithelial cells. Region I/lymphoid nuclear protein complexes (Nalm-6 [lane 14] and Raji [lane 16]) migrate slightly faster than region I/epithelial nuclear protein complexes (FHTE56 [lane 15] and Cal-1 [lane 17]).
As indicated, neither C/EBP or CTF/NF1 oligonucleotide probes inhibited the interaction between the region I probe and Nalm-6 nuclear protein (Fig 6A, lanes 3 and 4). In contrast, excess unlabeled double-stranded CBF/NF-Y markedly reduced region I binding to Nalm-6 nuclear protein (Fig 6A, lane 2). Excess unlabeled double-stranded CBF/NF-Y similarly reduced region I binding to FHTE56 nuclear protein (Fig 6B, lane 11). Taken together, these data strongly suggest that the region I binding factor is CBF/NF-Y. For this reason, additional supershift assays were performed in which Nalm-6 or FHTE56 nuclear protein was preincubated with CBF-A antisera42 before addition of 32P-labeled region I oligonucleotide probe (Fig 6B). As indicated, CBF-A antisera retarded the migration of the region I/nuclear protein complex, whereas an unrelated control (Oct-1) rabbit polyclonal antisera had no effect (Fig 6B, lanes 7 and 12 v lanes 8 and 13). These data further identify the CD10/NEP region I binding factor as CBF/NF-Y.
Tissue-Specific Expression of Alternatively Spliced CBF-B/NF-YA Subunits
The CBF/NF-Y transcription factor consists of three subunits, CBF-A, CBF-B, and CBF-C.42,47 The CBF-A and CBF-B subunits have been independently identified as NF-YB and NF-YA, respectively.43,44,48 The CBF-B/NF-YA subunit is alternatively spliced into long and short forms that contain or lack an exon encoding the majority of a glutamine-rich transactivation domain.44 Previous studies suggest that there is tissue-specific expression of the alternatively spliced CBF-B/NF-YA subunits, with primary expression of the long form in epithelial cells and the short form in lymphoid cells.44 For this reason, subtle differences in the migration of complexes containing CD10/NEP region I and nuclear protein from lymphoid and epithelial cells are of additional interest (Fig 6C). As indicated, complexes containing region I and nuclear protein from lymphoid cells (Nalm-6 and Raji) migrate slightly faster than complexes containing region I and nuclear protein from epithelial cells (FHTE56 and Calu-1) (Fig 6C, lanes 14 and 16 v lanes 15 and 17).
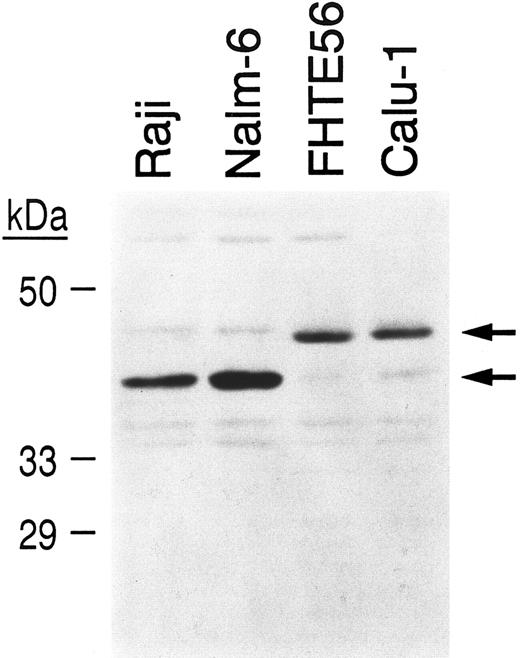
To determine whether tissue-specific differences in CBF-B/NF-YA subunit expression might explain differences in the mobility of complexes containing region I and lymphoid or epithelial nuclear protein (Fig 6C), nuclear extracts from Raji and Nalm-6 lymphoid cells and FHTE56 and Calu-1 epithelial cells were size-fractionated, blotted, and hybridized with an antibody directed against the CBF-B/NF-YA subunit43 (Fig 7). As indicated, Raji and Nalm-6 lymphoid cells primarily express the approximately 40-kD CBF-B/NF-YA short form, whereas FHTE56 and Calu-1 epithelial cells primarily express the 43-kD CBF-B/NF-YA long form (Fig 7).
Detection of alternatively spliced CBF-B/NF-YA subunits in lymphoid and epithelial cell extracts. Nuclear extracts from Raji and Nalm-6 lymphoid cells and FHTE56 and Calu-1 epithelial cells were size-fractionated, blotted, and incubated with an affinity-purified antiserum directed against the C-terminal peptide of NF-YA.35 The ∼40-kD CBF-B/NF-YA short form and the ∼43-kD CBF-B/NF-YA long form are indicated.
Detection of alternatively spliced CBF-B/NF-YA subunits in lymphoid and epithelial cell extracts. Nuclear extracts from Raji and Nalm-6 lymphoid cells and FHTE56 and Calu-1 epithelial cells were size-fractionated, blotted, and incubated with an affinity-purified antiserum directed against the C-terminal peptide of NF-YA.35 The ∼40-kD CBF-B/NF-YA short form and the ∼43-kD CBF-B/NF-YA long form are indicated.
Differential Effects of CBF/NF-YA Isoforms on CD10/NEP Region I Activity
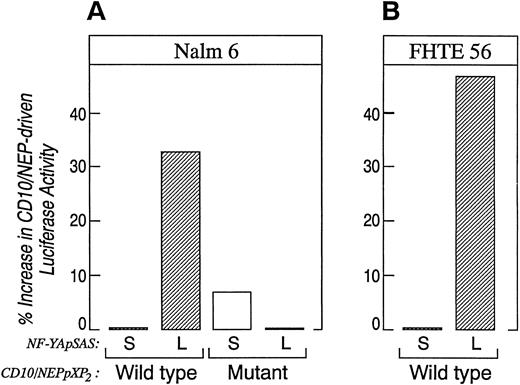
The data in Figs 6 and 7 suggest that tissue-specific differences in CD10/NEP region I activity might result from tissue-specific expression of alternatively spliced CBF-B/ NF-YA subunits. To explore this possibility, we directly compared the effects of the alternatively spliced CBF-B/NF-YA subunits on CD10/NEP-driven luciferase activity in Nalm-6 lymphoid cells. In initial experiments, Nalm-6 cells were cotransfected with CD10/NEP luciferase constructs containing an intact or mutated region I (CD10/NEP[−263/+147]pXP2 or I-M, respectively; Fig 5) and plasmids encoding the long or short forms of CBF-B/NF-YA (CBF-B/NF-YA short [S]pSAS or long [L]pSAS, respectively). CD10/NEP-driven luciferase activity was 33% higher in Nalm-6 cells cotransfected with CBF-B/NF-YA(L)pSAS and CD10/NEP(−263/+147)pXP2 than in cells cotransfected with CBF-B/NF-YA(S)pSAS and CD10/NEP(−263/+147)pXP2 (Fig 8A). However, CD10/NEP-driven luciferase activity in Nalm-6 cells cotransfected with CBF-B/NF-YA(L)pSAS and I-M was comparable to that in Nalm-6 cells cotransfected with either CBF-B/NF-YA(S)pSAS and I-M (CD10/NEP[−263/+147]pXP2) or CBF-B/NF-YA(S)pSAS and wild-type CD10/NEP(−263/+147)pXP2 . Taken together, these data indicate that adding the long form of CBF-B/NF-YA to cells that primarily express the short form of CBF-B/NF-YA specifically increases CD10/NEP region I promoter activity.
Comparison of CD10/NEP-driven luciferase activity in Nalm-6 and FHTE56 cells cotransfected the alternatively spliced CBF-B/NF-YA cDNAs. Nalm-6 cells (A) or FHTE56 cells (B) were cotransfected with 20 μg wild-type (W) or mutant (M) CD10/NEP (−263/+147) pXP2 , 40 μg of the CBF-B/NF-YA short pSAS (S) and CBF-B/NF-YA long pSAS (L), and 1 μg of CMV-GH. Luciferase activity was measured 48 hours posttransfection and normalized for GH production. Promoter activity (RLU/ng/mL) represents mean and standard errors for two separate experiments performed in duplicate or triplicate. Nalm 6 cells: W + S 37.1 ± 3.5 (baseline); W + L, 49.5 ± 5.7 (33% increase over baseline, P = .09); M + S 39.7 ± 3.6 (7% increase over baseline, P = NS); M + L 37.0 ± 3.7 (0% difference from baseline, P = NS). FHTE56 cells: W + S 69.1 ± 4.3 (baseline), W + L 101.9 ± 30.8 (47% increase over baseline, P = NS). The differences in CD10/NEP-driven luciferase activity in cells transfected with W + S, W + L, M + S, or M + L were evaluated using a one-sided Student's t-test.
Comparison of CD10/NEP-driven luciferase activity in Nalm-6 and FHTE56 cells cotransfected the alternatively spliced CBF-B/NF-YA cDNAs. Nalm-6 cells (A) or FHTE56 cells (B) were cotransfected with 20 μg wild-type (W) or mutant (M) CD10/NEP (−263/+147) pXP2 , 40 μg of the CBF-B/NF-YA short pSAS (S) and CBF-B/NF-YA long pSAS (L), and 1 μg of CMV-GH. Luciferase activity was measured 48 hours posttransfection and normalized for GH production. Promoter activity (RLU/ng/mL) represents mean and standard errors for two separate experiments performed in duplicate or triplicate. Nalm 6 cells: W + S 37.1 ± 3.5 (baseline); W + L, 49.5 ± 5.7 (33% increase over baseline, P = .09); M + S 39.7 ± 3.6 (7% increase over baseline, P = NS); M + L 37.0 ± 3.7 (0% difference from baseline, P = NS). FHTE56 cells: W + S 69.1 ± 4.3 (baseline), W + L 101.9 ± 30.8 (47% increase over baseline, P = NS). The differences in CD10/NEP-driven luciferase activity in cells transfected with W + S, W + L, M + S, or M + L were evaluated using a one-sided Student's t-test.
In additional experiments, FHTE56 cells were cotransfected with CD10/NEP (−263/+147) pXP2 and CBF-B/NF-YAshort(S)- or long(L)-pSAS (Fig 8B). CD10/NEP-driven luciferase activity was 47% higher in FHTE56 cells cotransfected with CBF-B/NF-YA(L)pSAS and CD10/NEP (263/+147) pXP2 than in cells cotransfected with CBF-B/NF-YA(S)pSAS and CD10/NEP (−263/+147) pXP2 . FHTE56 cells cotransfected with IM and either CBF-B/NF-YA(S)-pSAS or (L)-pSAS had significantly less CD10/NEP-driven luciferase activity than cells transfected with wild-type CD10/NEP (263/+147) pXP2 (data not shown). These data are consistent with the previous studies in which region I deletion or mutation decreased CD10/NEP-driven luciferase activity in FHTE56 cells (Figs 1 and 5). The additional cotransfections in FHTE56 cells further indicate that the short and long form of CBF-B/NF-YA have different effects on CD10/NEP region I promoter activity (Fig 8B).
DISCUSSION
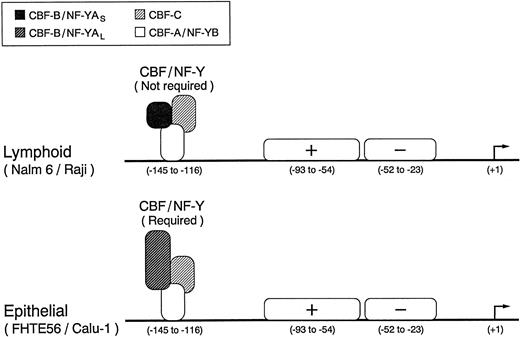
We have characterized the type 2 CD10/NEP major regulatory element and identified three functionally active transcription factor binding sites in regions I, II, and III. CBF/NF-Y binds to the inverted CCAAT box in region I, whereas a second positive and third negative factor bind to regions II and III, respectively (Fig 9). Although region I is required for maximal CD10/NEP-driven luciferase activity in the examined epithelial cell lines, this region is not required for maximal activity in the evaluated lymphoid cell lines (Fig 9). The apparent tissue-specific differences in the requirements for region I (and CBF/NF-Y) are of particular interest because lymphoid and epithelial cells express alternatively spliced versions of a CBF/NF-Y subunit that differ in biologic activity44 49 (Figs 7 and 8).
Identified regulatory elements in the type 2 CD10/NEP promoter. The three functionally active transcription factor binding sites in the type 2 CD10/NEP promoter are indicated. The heteromeric CBF/NF-Y complex, which includes the alternatively spliced CBF-B/NF-YA short or long form, binds to the inverted CCAAT box in region I, whereas a second positive and third negative factor bind to regions II and III, respectively. Although CBF/NF-Y appears to be required for maximal epithelial (FHTE56 and Calu-1) CD10/NEP type 2 promoter activity, the complex is not required for maximal lymphoid (Nalm-6 and Raji) CD10/NEP type 2 promoter activity.
Identified regulatory elements in the type 2 CD10/NEP promoter. The three functionally active transcription factor binding sites in the type 2 CD10/NEP promoter are indicated. The heteromeric CBF/NF-Y complex, which includes the alternatively spliced CBF-B/NF-YA short or long form, binds to the inverted CCAAT box in region I, whereas a second positive and third negative factor bind to regions II and III, respectively. Although CBF/NF-Y appears to be required for maximal epithelial (FHTE56 and Calu-1) CD10/NEP type 2 promoter activity, the complex is not required for maximal lymphoid (Nalm-6 and Raji) CD10/NEP type 2 promoter activity.
The heteromeric CBF/NF-Y transcription factor has an absolute requirement for the CCAAT motif and binds to CCAAT boxes in a variety of promoters including albumin, globin, β-actin, α-collagen, interleukin-4, and MHC class II genes.43,46,50-52 As is frequently the case, the CCAAT element in the CD10/NEP promoter is inverted and included in an 11-bp CBF/NF-Y consensus sequence.28 The CBF/NF-Y complex consists of three subunits that have been separately described as CBF-A/NFY-B, CBF-B/NF-YA, and CBF-C.47,48,53,54 The CBF/NF-Y subunits contain no known protein-protein interaction motifs, suggesting that CBF/NF-Y may be a unique heteromeric DNA binding protein.54
The CBF-B/NF-YA and CBF-A/NF-YB subunits are highly conserved through evolution and functionally interchangeable with their yeast homologs, HAP2 and HAP3.48,55,56 The homology between HAP2 and CBF-B/NF-YA led to the identification of DNA-binding and subunit interaction motifs in the CBF-B/NF-YA carboxy terminus.54 However, the alternatively spliced sequences encoding the glutamine-rich domain of CBF-B/NF-YA are located in the amino terminus.44 Therefore, the manner in which tissue-specific expression of CBF-B/NF-YA short and long forms affects CD10/NEP region I activity remains to be determined. Recent analyses suggest that there are at least three discrete interactions involving (1) the CBF-C and CBF-A/NF-YB subunits, (2) the heterodimer consisting of CBF-A/NF-YB and CBF-C and the additional CBF-B/NF-YA subunit, and (3) the CBF/NF-Y heterotrimer and DNA.42,47 57 The demonstrated differences in the biologic activity of CBF/NF-Y complexes containing the long or short CBF-B/NF-YA subunits may be further explored using the region I CD10/NEP luciferase constructs (Figs 5 and 8).
In previous analyses, CBF/NF-Y was shown to interact with and stabilize additional upstream transcription factors via protein-protein interactions.58 In this regard, it is noteworthy that CBF/NF-Y interacts with upstream SP-1 factors,59 and that the upstream type 2 CD10/NEP sequence is GC-rich and contains potential SP-1 binding sites.32 Although CBF/NF-Y is known to interact with upstream factors, there are no documented examples of CBF/NF-Y interaction with downstream factors. This is of interest because CD10/NEP-driven luciferase activity is similar in lymphoid cells transfected with CD10/NEPpXP2 constructs containing intact or mutated CBF/NF-Y binding sites (Fig 5). Therefore, downstream transcription factors binding to regions II and III of the CD10/NEP promoter are less likely to require CBF/NF-Y interactions for stability and/or activity. Furthermore, the fact that a mutated CBF/NF-Y binding site reduced CD10/NEP-driven luciferase activity in epithelial cells (Fig 5) suggests that the additional region II and III binding factors are insufficient for maximal CD10/NEP activity in these cells. Additional characterization of CBF/NF-Y subunit interactions in the CD10/NEP promoter and identification of region II and III binding factors are likely to provide further insight into the regulation of this important enzyme.
ACKNOWLEDGMENT
We gratefully acknowledge B. de Crombrugghe for the gift of the anti–CBF-A antibody and R. Mantovani for the gift of the NF-YA isoform cDNAs in eukaryotic expression vectors and the NF-YA antisera. We thank D. Tenen and D.E. Zhang for their helpful comments throughout this project, and Donna Favreau for manuscript preparation.
Supported by Public Health Service Grant No. CA-55095 from the National Cancer Institute.
Address reprint requests to Margaret A. Shipp, MD, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115.

![Fig. 4. EMSA of the type 2 CD10/NEP promoter with mutated region I, II, or III probes. (A) Sequences of wild-type and mutant region I, II, or III oligonucleotide probes. The 10-bp mutations in region I, II, and III oligonucleotide probes are indicated. (B, C, and D) EMSAs. A double-stranded oligonucleotide of wild-type (W) region I (B), region II (C), or region III (D) was 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 in the absence (−, lanes 1, 5, and 9) or presence of 100-fold molar excess unlabeled W (lanes 2, 6, and 10) or mutated ([M] lanes 3, 7, and 11) oligonucleotide. Double-stranded mutated region I, II, or III oligonucleotide probes (M) were also 32P-labeled and incubated with 5 μg nuclear protein from Nalm-6 (lanes 4, 8, and 12). Arrows identify the major shifted bands.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/11/10.1182_blood.v89.11.4136/4/m_bl_0026f4.jpeg?Expires=1767823114&Signature=udWsfS3XVkh3QS~jR5YW--7-TlnRqQR0N4ZymmRjMwQRlDOb~mJdJdF~-rfnwFlQYXKya4Rqf3EIr2JW9qLmrvxdkxaAfJ4SXvz-tvGo4SYSONjn6tjb0zRLDEdypYBRIxRb3DSQ0cOj-uZMvWwyfKljyOUYmDINDXlooLqHW1cCwLhpEzVY63mJ-Ny3sLzknHJo1FY4psuD1zl5Ur4pU1etF4ZYsl2u-R9mZg4zARjLx5ijAbtncWYhWMugJmKMH-V8VJe~rrW94OjVUKzcz1EQblyTBVCFUXgHn9F7IMs0rmJGLtXY~WnyDjdBFYRTuG7S12hlufd1b~RqdMz0~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. (A) EMSAs of CD10/NEP region I in the presence or absence of oligonucleotides from known CCAAT binding factors. A double-stranded region I oligonucleotide was 32P-labeled and incubated with 5 μg Nalm-6 nuclear protein in the absence (−) (lane 1) or presence of 100-fold molar excess of the indicated unlabeled double-stranded oligonucleotides: CBF/NF-Y, lane 2; C/EBP, lane 3; CTF/NFI, lane 4; self, lane 5. (B) EMSAs of CD10/NEP region I in the presence or absence of a CBF/NF-Y oligonucleotide or antibody. 32P-labeled double-stranded region I oligonucleotide was also incubated with Nalm-6 nuclear protein in the absence (−) (lane 6) or presence of anti–CBF-A [NF-Y] antibody (lane 7) or anti-OCT1 antibody (lane 8), or with FHTE56 nuclear protein in the absence (−) (lane 9) or presence of unlabeled self-oligonucleotide probe (lane 10), unlabeled CBF/NF-Y oligonucleotide probe (lane 11), anti–CBF-A [NF-Y] antibody (lane 12), or anti-Oct1 antibody (lane 13). Upper arrow indicates supershifted bands and lower arrow indicates major shifted bands in the absence of specific antibodies. (C) Electrophoretic mobility of region I/nuclear protein complexes from lymphoid and epithelial cells. Region I/lymphoid nuclear protein complexes (Nalm-6 [lane 14] and Raji [lane 16]) migrate slightly faster than region I/epithelial nuclear protein complexes (FHTE56 [lane 15] and Cal-1 [lane 17]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/89/11/10.1182_blood.v89.11.4136/4/m_bl_0026f6.jpeg?Expires=1767823114&Signature=l5MROkVR1xwKf0WKEOMdGnu6lmLEbGPmUEum5wz7oAc5zv3IqRta--kTEP9-~hzBUcZsD1UveFr4J9Xxf9-I5gKdbJdRNzvlvMs7tMbmUwLAIuEoNJ-92JlcZ-2vc7nFKX8v~DXtdNqGLBcjRMT4Yf0PeV0UNFbiVWL-3dedQTN3cRu9KkaAolv27EzCunzeOiOrQ5nSKrG9dhq0usP-9y4LVy51-OdQbV5ghITwWH-fdkbaTs60gPTguamqmDgnUOOpD-tccm335myyUm65Aytkhs7mjtu-8PGAQ776ZqiNCMy5iJcCzWdQ7WHqpDid8xgeKYnOeALLbI7IB0f3zA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal