Abstract
Genes of the major histocompatibility complex (MHC) are associated with susceptibility to different immune and nonimmune mediated diseases. We had reported that the drug adverse reaction, clozapine-induced agranulocytosis (CA), is associated with different HLA types and HSP70 variants in Ashkenazi Jewish and non-Jewish patients, suggesting that a gene within the MHC region is associated with CA. This study was designed to find common genetic markers for this disorder in both ethnic groups. The tumor necrosis factor (TNF ) microsatellites d3 and b4 were found in higher frequencies in both Jewish and nonJewish patients: 51 of 66 (77%) and 48 of 66 (57%), respectively. Comparisons of these frequencies with those of controls, 28 of 66 (42%) and 18 of 66 (27%), were statistically significant (corrected P value = .001 for the d3 allele and .0005 for the b4 allele). On the other hand, the TNF microsatellite b5 was underrepresented in the group of patients, 9 of 66 (14%), when compared with the control subjects, 43 of 66 (65%) (corrected P value = .0005), probably related to protection from CA. Our results show a strong association of some genetic variants of the TNF loci with susceptibility to CA in two different ethnic groups suggesting involvement of TNF and/or associated gene(s) products in the pathogenesis of this hematologic-drug adverse reaction.
CLOZAPINE (Clozaril; Sandoz Inc, Berne, Switzerland) is a new antipsychotic drug with absolute indications for the treatment of drug-resistant schizophrenia and of patients who are unable to tolerate traditional antipsychotic medications.1 Additionally, clozapine is considered more efficient than traditional antipsychotic drugs and may also be useful in the treatment of other neurologic disorders.1 However, its use has been limited mainly because of the idiosyncratic drug-induced agranulocytosis that occurs in approximately 1% of the population treated with the drug.2,3 Susceptibility to this life-threatening disorder is linked to genetic factors, including: (1) HLA-DR4 and B38 in Ashkenazi Jewish patients and (2) HLA-DR2 in non-Jewish patients.4 Additionally, the high incidence of recurrence of agranulocytosis in patients rechallenged with the drug points toward a genetic predisposition in the pathogenesis of this disorder.5 Different mechanisms are probably involved, but to date there is no clear evidence of an immune or directly toxic effect of the drug or its metabolites on cells of the myeloid lineage.6-13
Studies of the HLA alleles located in the short arm of chromosome 6, region 6p21, have been useful in understanding immune responses, transplantation, and forensic science.14,15HLA alleles or haplotypes are associated with several immune- and nonimmune-mediated diseases as well as with adverse drug reactions to xenobiotics.16-18 At least one third of haplotypes of unrelated individuals are the product of nonrandom association of alleles of the major histocompatibility complex (MHC) loci. These have been named extended haplotypes and implies that they carry specific alleles in a region defined by their HLA-B, complotype, and DR/DQ variants. Unrelated individuals with the same extended haplotypes are expected to have common MHC alleles.15,19 20
It is not clear why different alleles are associated with clozapine-induced agranulocytosis (CA) in Jewish and non-Jewish patient populations. It is possible that an abnormal gene product that mediates agranulocytosis is encoded within the HLA-B, DR region, in the intermediate region of the MHC, would be common to patients of several ethnic groups. The association of CA with different HLA types in Jewish and non-Jewish patients with CA could therefore be due to linkage disequilibrium of a common marker for both groups within the MHC region. In the absence of complotypes and family studies, it is possible to assign known nonrandom associated haplotypes with high delta value that represent linkage disequilibrium.21 In previous studies we described findings consistent with the hypothesis that a dominant gene within the MHC region, marked by HSP70 variants, is associated with CA in the two different ethnic groups studied.22 These findings suggested that a second candidate explanation of the MHC associations are the tumor necrosis factor (TNF ) genes. These genes [TNF-α, TNF-β (LTα), and LTβ] are located in a 7-kb span of genomic DNA in the class-III region.15 They are marked by variants that have shown linkage disequilibrium with HLA-B and DR alleles.23 24
Therefore, we have studied the frequencies of the polymorphisms (four dinucleotide microsatellites and two single base variants) of the TNF variants in the intermediate region of the MHC in patients with CA from two different ethnic backgrounds.
MATERIALS AND METHODS
Patients
We arranged with the Clozaril Monitoring System to notify us of nationwide agranulocytosis cases and to identify the attending physicians.25 To maintain confidentiality, patient identities were not disclosed. Clinical case information was reviewed to determine the patient's suitability for the study's criteria for agranulocytosis, ie, an absolute neutrophil count less than 500/μL in the course of treatment by clozapine. The treating doctors of patients meeting these criteria were sent letters explaining the purpose and nature of the study and requesting assistance in facilitating access to the patient for the purpose of obtaining blood samples. With a patient's consent, additional information was obtained about the case history of the disease, including indication for clozapine, duration and dose of clozapine treatment, concomitant medication, and medical history. A total of 33 schizophrenic patients (12 Jewish and 21 non-Jewish) who had agranulocytosis during treatment with clozapine were included in the study. Ashkenazi Jewish or non-Jewish ethnicity was determined by historical evidence of the patient's four grandparents. Patients were not related; the patients and controls included in this study were not from inbred communities. All patients were white and of European origin. The patients included in this study did not have relatives available at the time of the collection of blood samples, and therefore phenotypes only were ascertained. The mean age of the patients who developed agranulocytosis was 39 years and the mean age in the group of controls was 33 years (P corrected [Pc] = not significant). The percentage of men was 83% (10 of 12) in the Ashkenazi Jewish group of patients and 38% (8 of 21) in the non-Jewish group of patients. Overall, the sex distribution in the group of patients was 65% (18 of 33) men and 35% (15 of 33) women.
Controls
The control group consisted of 33 genetically unrelated, white schizophrenic patients of European ancestry (18 Ashkenazi Jewish and 15 non-Jewish) who were treated with clozapine for at least 52 weeks but did not develop agranulocytosis. Control patients were ascertained from the clinical services of Hillside Hospital, Long Island Jewish Medical Center (Long Island, NY), where they were receiving treatment with clozapine. The clozapine dose was titrated gradually, on the basis of clinical judgment, to the level that produced the best clinical response. No difference was found in the dose administered to the patients who developed agranulocytosis and those who did not.25 Because all patients (agranulocytosis and controls) received clozapine in the context of the Clozaril Monitoring System (white blood cell counts [WBCs]), they underwent clinical and hematologic assessment on a weekly basis.25 Similarly, probands and controls had treatment-resistant schizophrenia, intolerant schizophrenia, or schizoaffective disorder, which was a requirement for eligibility for clozapine treatment. Thus, insofar as frequency of assessment and form of illness, probands and control groups did not differ. There were 78% (14 of 18) males in the Ashkenazi Jewish control group and 73% (11 of 15) males in the non-Jewish control population. Overall, the sex distribution in the control group was 76% (25 of 33) males and 24% (8 of 33) females. All these subjects had previously been typed for class I and II HLA alleles.4
TNF Constellation Polymorphism Typing
All patients and controls had previously been typed for class I and II HLA alleles,4 and were also typed for the TNF polymorphisms as described before.23,24 26-28
TNF microsatellites a-b, d-e. The primers and polymerase chain reaction (PCR) conditions for the TNF microsatellite typing were those reported previously in the literature.23 For the first round of amplification, different primer combinations were used. For the TNF microsatellites a-b, the primers were sense: 5′ GCCTCTAGATTTCATCCAGCCACA 3′ and antisense: 5′ CCTCTCTCCCCTGCAACACACA 3′. For the TNF microsatellites d-e, the primers were sense: 5′ AGATCCTTCCCTGTGAGTTCTGCT 3′ and antisense: 5′ TGAGACAGAGGATAGGAGAGACAG 3′. For these amplifications, 200 ng of genomic DNA were processed in a 20-μL final reaction volume containing 2 μL of 10× PCR buffer, 2 μL of 10× dNTPs mix, 1 μL of each primer (10 pmol/L stock) and 0.2 μL of Taq polymerase (Perkin-Elmer Cetus, Cambridge, MA). The PCR reaction was performed in a thermal cycler (9600, Perkin-Elmer Cetus) under the following conditions: 3 minutes at 94°C, followed by 35 cycles of 1 minute at 94°C, 1 minute at 60°C, 1 minute at 72°C, followed by a final extension for 10 minutes at 72°C. After the first PCR amplification, an aliquot (2 μL), was used as a template for the second PCR amplification round using the “heminested” PCR strategy with the following primers: for TNFa, antisense: 5′ GCACTCCAGCCTAGGGAGA 3′; for TNFb, sense: 5′ GTGTGTGTTGCAGGGGAGAGAG 3′; for TNFd, antisense: 5′ CATAGTGGGACTCTGTCTCCAAAG 3′; and for TNFe, sense: 5′ GTGCCTGGTTCTGGAGCCTCTC 3′ using conditions as described for the first PCR amplification but using 5 cycles with 0.1 μL of α32P dCTP per each PCR reaction. After the final PCR amplification, 5 μL of the PCR products were added to 5 μL of a formamide-containing stop solution (US Biochemicals, Cleveland, OH) heated at 94°C for 2 minutes and analyzed by electrophoresis in a 0.4-mm thick, 6% polyacrylamide sequencing gel (GIBCO-BRL Life Technologies, Gaithersburg, MD) at 30 W for 4 hours. For autoradiography, the gels were dried and exposed for 2 to 12 hours using Kodak X-OMAT film (Eastman Kodak, Rochester, NY) at −70°C with an intensifying screen.
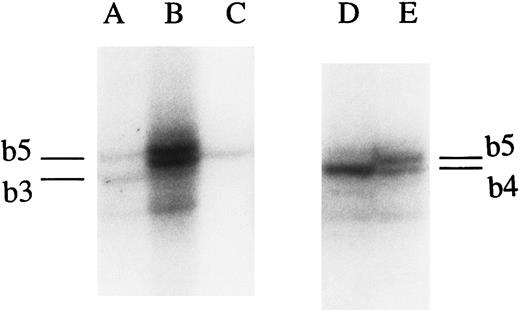
The microsatellites were assigned using reference homozygous cells (Tenth International Histocompatibility Workshop),28a as follows: for TNFa1, BTB; for TNFa2, VAVY; for TNFa3, COX; for TNFa4, SLE005; for TNFa5, TISI; for TNFa6, HOM2; for TNFa7, LBUF; for TNFa8, MOU; for TNFa10, BM16; for TNFa11, HHKB; for TNFa12, TUBO; for TNFb1, TUBO; for TNFb3, VAVY; for TNFb4, LBUF; for TNFb5, TISI; for TNFd1, VAVY; for TNFd3, BTB; for TNFd4, JHAF; for TNFd5, TISI; for TNFd7, TUBO 89; for TNFe1, LWAGS; and for TNFe3, LBUF. In addition, several heterozygous samples were typed in the CA population. To demonstrate that it is possible to distinguish microsatellite variants, including those that have close electrophoretic migration, an example is shown in Fig 1. The panel of homozygous cell lines with known TNF microsatellite alleles had been characterized in the past by other investigators23 24 and was confirmed by our experiments. New information added in these studies was the assignment of variants of the TNFα(-308) A/G polymorphism (see Table 1).
Autoradiography of the electrophoresis of PCR products after amplification of CA samples. A 6% polyacrylamide gel was used as described in Materials and Methods. (A) Sample heterozygous for alleles b3 and b5. (B) Sample heterozygous for the b4 and b5 alleles. (C) Sample homozygous for the b4 allele. (D) Sample homozygous for the b4 allele. (E) Sample heterozygous for the b4 and b5 alleles.
Autoradiography of the electrophoresis of PCR products after amplification of CA samples. A 6% polyacrylamide gel was used as described in Materials and Methods. (A) Sample heterozygous for alleles b3 and b5. (B) Sample heterozygous for the b4 and b5 alleles. (C) Sample homozygous for the b4 allele. (D) Sample homozygous for the b4 allele. (E) Sample heterozygous for the b4 and b5 alleles.
TNF Gene Constellation in White Extended Haplotypes
| HLA-DR . | Complotype . | HSP70 −2, −1 . | TNF . | HLA-B . | Haplotype No. . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | C4B . | C4A . | BF . | C2 . | . | e . | d . | α-308 . | βn . | a . | b . | . | . |
| 3 | 1 | 0 | S | C | 8.5 − C | 3 | 1 | 2 | 1 | 2 | 3 | 8 | 10 |
| 2 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 11 | 4 | 7 | 12 |
| 7 | 1 | 3 | F | C | 9 − A | 3 | 3 | 1 | 2 | 7, 8 | 4 | 44 | 4 |
| 4 | 0 | 3 | S | C | 9 − A | 3 | 3 | 1 | 1 | 6, 7 | 5 | 44 | 4 |
| 7 | 1 | 6 | S | C | 9 − A | 3 | 4 | 1 | 2 | 2 | 5 | 57 | 4 |
| 5 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 5 | 5 | 35 | 4 |
| 1 | 1, 2 | 2 | S | C | 9 − A | 1 | 4 | 1 | 2 | 2 | 1 | 14 | 4 |
| 4 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 10 | 4 | 35 | 3 |
| 4 | 1 | 2 | S | C | 9 − A | 3 | 3 | 1 | 2 | 10 | 4 | 38 | 10 |
| 1 | 0 | 3, 2 | F | C | 9 − A | 1 | 4 | 1 | 2 | 5 | 5 | 35 | 1 |
| 4 | 3 | 3 | S | C | 9 − A | 1 | 4 | 1 | 2 | 2 | 1 | 62 | 4 |
| 2 | 2 | 4 | S | 0 | 9 − A | 3 | 3 | 1 | 2 | 10 | 4 | 18 | 2 |
| 3 | 0 | 3 | F1 | C | 8.5 − C | 3 | 4 | 1 | 2 | 1 | 5 | 18 | 8 |
| 7 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 7 | 4 | 13 | 2 |
| HLA-DR . | Complotype . | HSP70 −2, −1 . | TNF . | HLA-B . | Haplotype No. . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | C4B . | C4A . | BF . | C2 . | . | e . | d . | α-308 . | βn . | a . | b . | . | . |
| 3 | 1 | 0 | S | C | 8.5 − C | 3 | 1 | 2 | 1 | 2 | 3 | 8 | 10 |
| 2 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 11 | 4 | 7 | 12 |
| 7 | 1 | 3 | F | C | 9 − A | 3 | 3 | 1 | 2 | 7, 8 | 4 | 44 | 4 |
| 4 | 0 | 3 | S | C | 9 − A | 3 | 3 | 1 | 1 | 6, 7 | 5 | 44 | 4 |
| 7 | 1 | 6 | S | C | 9 − A | 3 | 4 | 1 | 2 | 2 | 5 | 57 | 4 |
| 5 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 5 | 5 | 35 | 4 |
| 1 | 1, 2 | 2 | S | C | 9 − A | 1 | 4 | 1 | 2 | 2 | 1 | 14 | 4 |
| 4 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 10 | 4 | 35 | 3 |
| 4 | 1 | 2 | S | C | 9 − A | 3 | 3 | 1 | 2 | 10 | 4 | 38 | 10 |
| 1 | 0 | 3, 2 | F | C | 9 − A | 1 | 4 | 1 | 2 | 5 | 5 | 35 | 1 |
| 4 | 3 | 3 | S | C | 9 − A | 1 | 4 | 1 | 2 | 2 | 1 | 62 | 4 |
| 2 | 2 | 4 | S | 0 | 9 − A | 3 | 3 | 1 | 2 | 10 | 4 | 18 | 2 |
| 3 | 0 | 3 | F1 | C | 8.5 − C | 3 | 4 | 1 | 2 | 1 | 5 | 18 | 8 |
| 7 | 1 | 3 | S | C | 9 − A | 3 | 3 | 1 | 2 | 7 | 4 | 13 | 2 |
TNF A/G single base variants polymorphisms (α-308, βn).The A/G single-base polymorphisms in the TNFα promoter (-308) and the TNFβ second intron were analyzed by the PCR-restriction fragment length polymorphism method using the Nco I restriction enzyme as described before.26,28 The primers used for the analysis of the TNFα promoter (-308) A/G polymorphism were sense: 5′ AGGCAATAGGTTTTGAGGGCCAT 3′ and antisense: 5′ TCCTCCCTGCTCCGATTCCG 3′. The primers used for the analysis of the TNFβ second intron (A/G) polymorphism were sense: 5′ CCGTGCTTCGTGCTTTGGACTA 3′ and antisense: 5′ AGAGCTGGTGGGACATGTCTG 3′. For these amplifications, 200 ng of genomic DNA were added to 20 μL of PCR reaction mixture containing 0.5 pmol/L of each primer, 0.25 mmol/L of each dNTP, 1.5 mmol/L MgCl2 and 1 U of Taq polymerase (Perkin-Elmer Cetus). The cycling conditions for these amplifications were as follows: 1 cycle of 94°C for 3 minutes, 60°C for 1 minute, 72°C for 1 minute; 35 cycles of 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute; 1 cycle of 94°C for 1 minute, 60°C for 1 minute, 72°C for 5 minutes. Restriction digests of the PCR products were generated in a 25-μL final volume using 10 to 20 U of the restriction enzyme Nco I (GIBCO-BRL, New York, NY) for 16 to 24 hours. The TNFα promoter (-308) A/G restriction digests generated products of 87 and 20 bp for allele *α1 and of 107 bp for allele *α2, and were analyzed on a 4% agarose gel (2% NuSieve, 2% GIBCO-BRL). The TNFβ second intron (A/G) restriction digests generated a 740-bp fragment for the allele *β1 and a 555-bp plus 185-bp fragment for the allele *β2, and were analyzed on a 1% agarose gel (GIBCO-BRL).26 28
Statistical Analysis
All data analysis was performed with the aid of the Instat software system (Graphpad, San Diego, CA). P and odds ratio values were determined by the Fisher's exact test and all P values were corrected for the number of comparisons to obtain the Pc value.18
RESULTS
The data demonstrating significant association of HLA class II alleles with CA are summarized in Table 2. This table shows that in Ashkenazi Jewish patients, HLA-DRB1*0402, HLA-DQB1*0302, and HLA-DQA1*0301 independently or together, presumably as a haplotype, were increased in patients with CA when compared with those without it. In non-Jewish patients, the HLA-DRB1*02, DQB1*0502, and DQA1*0102 alleles, independently or together, presumably as a haplotype, were increased in patients with CA when compared with those without it.
Associations of TNF Variants, MHC Class II Alleles, and MHC Haplotypes in Patients With CA
| . | Allele or Haplotype . | Genotype Frequencies . | Pc Value . | |
|---|---|---|---|---|
| . | . | CA* Patients . | Controls . | . |
| Ashkenazi | HLA-DRB1*0402 | 11/24 | 6/54 | NS† |
| Jewish | HLA-DQB1*0302 | 11/24 | 8/54 | NS† |
| HLA-DQA1*0301 | 12/24 | 13/54 | NS† | |
| TNFd3 | 17/24 | 11/36 | .02 | |
| TNFb4 | 19/24 | 11/36 | .002 | |
| TNFb5 | 1/24 | 21/36 | .0005 | |
| [HLA-DRB1*0402, DRB4*0101, DQB1*0302, DQA1*0301, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa10, TNFb4, HLA-B38 ] | 12/24 | 3/36 | NS† | |
| Non-Jewish | HLA-DRB1*02 | 14/40 | 4/32 | NS† |
| HLA-DQB1*0502 | 10/40 | 1/32 | NS† | |
| HLA-DQA1*0102 | 15/40 | 3/32 | .04 | |
| TNFd3 | 34/42 | 17/30 | NS | |
| TNFb4 | 29/42 | 6/30 | .0005 | |
| TNFb5 | 8/42 | 22/30 | .0005 | |
| [HLA-DRB1*02+, DRB5*02, DQB1*0502, DQA1*0102, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa11, TNFb4] | 10/40 | 0/32 | NS† | |
| Combined | TNFd3 | 51/66 | 28/66 | .001 |
| TNFb4 | 48/66 | 17/66 | .0005 | |
| TNFb5 | 9/66 | 43/66 | .0005 | |
| [HLA-DRB1*0402, DRB4*0101, DQB1*0302, DQA1*0301, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa10, TNFb4, HLA-B38 ] | 12/64 | 4/66 | NS | |
| [HLA-DRB1*02 #, DRB5*02, DQB1*0502, DQA1*0102, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa11, TNFb4] | 10/64 | 0/66 | NS | |
| . | Allele or Haplotype . | Genotype Frequencies . | Pc Value . | |
|---|---|---|---|---|
| . | . | CA* Patients . | Controls . | . |
| Ashkenazi | HLA-DRB1*0402 | 11/24 | 6/54 | NS† |
| Jewish | HLA-DQB1*0302 | 11/24 | 8/54 | NS† |
| HLA-DQA1*0301 | 12/24 | 13/54 | NS† | |
| TNFd3 | 17/24 | 11/36 | .02 | |
| TNFb4 | 19/24 | 11/36 | .002 | |
| TNFb5 | 1/24 | 21/36 | .0005 | |
| [HLA-DRB1*0402, DRB4*0101, DQB1*0302, DQA1*0301, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa10, TNFb4, HLA-B38 ] | 12/24 | 3/36 | NS† | |
| Non-Jewish | HLA-DRB1*02 | 14/40 | 4/32 | NS† |
| HLA-DQB1*0502 | 10/40 | 1/32 | NS† | |
| HLA-DQA1*0102 | 15/40 | 3/32 | .04 | |
| TNFd3 | 34/42 | 17/30 | NS | |
| TNFb4 | 29/42 | 6/30 | .0005 | |
| TNFb5 | 8/42 | 22/30 | .0005 | |
| [HLA-DRB1*02+, DRB5*02, DQB1*0502, DQA1*0102, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa11, TNFb4] | 10/40 | 0/32 | NS† | |
| Combined | TNFd3 | 51/66 | 28/66 | .001 |
| TNFb4 | 48/66 | 17/66 | .0005 | |
| TNFb5 | 9/66 | 43/66 | .0005 | |
| [HLA-DRB1*0402, DRB4*0101, DQB1*0302, DQA1*0301, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa10, TNFb4, HLA-B38 ] | 12/64 | 4/66 | NS | |
| [HLA-DRB1*02 #, DRB5*02, DQB1*0502, DQA1*0102, HSP70-2*A, HSP70-1*9, TNFe3, TNFd3, TNFα(−308)*1, TNFβn(A/G)*2, TNFa11, TNFb4] | 10/64 | 0/66 | NS | |
The P values not corrected were as follows: Jewish: .02 for DRB1*0402, 0.008 for DQB1*0302, 0.003 for DQA1*0301, .02 for the haplotype; Non-Jewish: .03 for DRB1*02, .02 for DQB1*0502, .007 for DQA1*0102, .002 for the haplotype.
Abbreviations: NS, not significant; +, can be either *1501 or *1601.
Clozapine-induced agranulocytosis.
These P values were significant because they confirmed previous hypothesis based on pilot studies.45
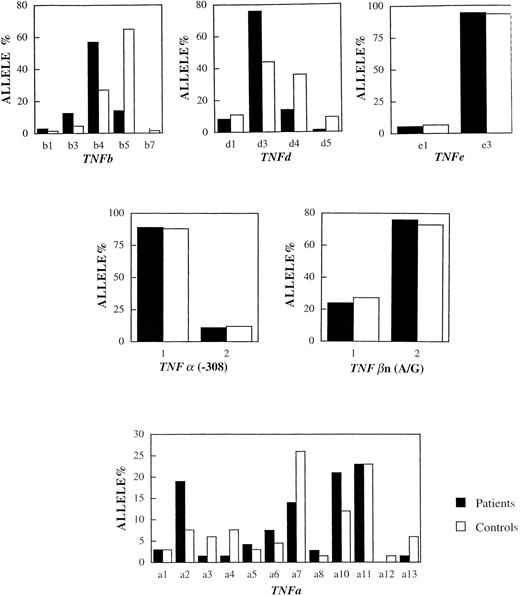
Table 2 shows the frequencies of the TNF polymorphisms in the 33 patients and 33 controls. In the group of patients, the frequencies of the d3 and b4 alleles were 77% and 72%. In the control group, the same alleles were present with frequencies of 42% and 27%. The frequencies of the d3 and b4 alleles in CA patients were significantly higher than those of the controls, for the b4 marker (odds ratio ∼8, Pc = .0005) and for the d3 marker (odds ratio ∼4, Pc = .001). On the other hand, the microsatellite allele b5 was underrepresented in the patients, 9 of 66 patient alleles (14%) against 43 of 66 control alleles (65%). This difference was statistically significant (Pc = .0005). There was no significant difference in the frequency of the TNFa and TNFe microsatellite alleles or the TNFα(-308) and TNFβn(A/G) polymorphisms between patients and controls. It is clear that the TNF variants have different frequencies in CA patients and controls, independent of the ethnic origin of the patients and controls tested (see Fig 2).
The distribution of (microsatellite) alleles in CA patients versus control individuals. Horizontal axis shows the number (n) of tandem repeats. Vertical axis shows the frequency of the alleles in the populations studied. The frequencies of the d3 and b4 alleles were significantly higher in the patients whereas the frequency of the b5 allele was significantly higher in the controls (see text for details).
The distribution of (microsatellite) alleles in CA patients versus control individuals. Horizontal axis shows the number (n) of tandem repeats. Vertical axis shows the frequency of the alleles in the populations studied. The frequencies of the d3 and b4 alleles were significantly higher in the patients whereas the frequency of the b5 allele was significantly higher in the controls (see text for details).
The microsatellites TNFa, b, d, e, as well as the TNFα promoter (-308) A/G and TNFβ second intron (A/G) polymorphisms were studied in a panel of homozygous cells previously typed for HLA-B, HLA-DR, HSP70-2, -1, and complotype. The homozygous cells used had been previously reported from family studies corresponding to individuals homozygous for extended haplotypes.15 The assignments of variants of microsatellites TNFa, b, and c to extended haplotypes has been described before.23,24 Table 1 shows assignments of HLA-DR, complotype, HSP70-1, -2; TNF constellation and HLA-B in 14 white extended haplotypes. There are 7 of the 14 extended haplotypes listed with unique and different TNF constellations. The remaining 7 are marked by shared TNF constellations; B38, DR4 and B35, DR4 share the TNFa10, b4, βn2, α(-308)1, d3, e3, B35, DR1 and B62, DR4 share TNFa2, b1, βn2, α(-308)1, e1, d4. Also, the extended haplotypes marked by B44 can be distinguished by their TNF polymorphisms. However, the HLA-B44, DR7 haplotype shares a TNF constellation with HLA-B13, DR7 haplotype. Of interest, the known association of HLA-B38, TNFb4, a10, βn2, α (-308)1, e3, d3 with DRB1*0402 was useful in assigning haplotypes in CA patients of Jewish ancestry as shown in Table 3.23,24 Additionally, the B7, DR2 haplotype and the DR2 carrying haplotypes associated with TNF b4, a11, βn2, α(-308)1, e3, d3, DRB1*1501 were useful to a lesser degree in non-Jewish CA patients as shown in Table 4. The assignment of high delta haplotypes was based on well-known nonrandom association of HLA specificities and alleles.21 It is clear that the haplotype assignment was easier in Jewish patients. In non-Jewish patients it was possible in 10 of 21 patients. In addition, 7 of 8 patients with DRB1*1601 were assumed to carry TNF constellations marked by TNFa10, b4 or TNFa11, b4. Therefore, the overrepresentation of the TNFb4 and d3 alleles is because of the presence of haplotypes carrying B44, DR7, or DR2 in non-Jewish patients and B38, DR4 in Jewish patients as shown in Fig 3.
TNF Constellation Polymorphism in Jewish CA Patients
| First Haplotype . | Second Haplotype . |
|---|---|
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B38 | HLA-DRB1*0701 (DR7), TNFe3, d7, α(−308)1, βn2, a7, b4, HLA-B44 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B38 | HLA-BRB1*0701 (DR7), TNFe1, d3, α(−308)1, βn2, a11, b4, HLA-B37 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a7, b4, HLA-B44 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a7, b4, HLA-B55 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1502 (DR2), TNFe3, d3, α(−308)1, βn2, a13, b5, HLA-B5 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1001 (DR10), TNFe3, d3, α(−308)1, βn2, a2, b5, HLA-Bx |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1401 (DR6), TNFe3, d5, α(−308)1, βn2, a2, b5, HLA-B44 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1401 (DR6), TNFe3, d5, α(−308)1, βn2, a7, b1, HLA-B41 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a2, b3, HLA-B8 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*1301 (DR6), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B45 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| First Haplotype . | Second Haplotype . |
|---|---|
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B38 | HLA-DRB1*0701 (DR7), TNFe3, d7, α(−308)1, βn2, a7, b4, HLA-B44 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B38 | HLA-BRB1*0701 (DR7), TNFe1, d3, α(−308)1, βn2, a11, b4, HLA-B37 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a7, b4, HLA-B44 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a7, b4, HLA-B55 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1502 (DR2), TNFe3, d3, α(−308)1, βn2, a13, b5, HLA-B5 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1001 (DR10), TNFe3, d3, α(−308)1, βn2, a2, b5, HLA-Bx |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1401 (DR6), TNFe3, d5, α(−308)1, βn2, a2, b5, HLA-B44 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1401 (DR6), TNFe3, d5, α(−308)1, βn2, a7, b1, HLA-B41 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a2, b3, HLA-B8 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*0402 (DR4), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B38 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*1301 (DR6), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B45 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
Haplotypes or generic types with high delta values are in italics. Statistically significant alleles are written in bold.
TNF Constellation Polymorphism in Non-Jewish CA Patients
| First Haplotype . | Second Haplotype . |
|---|---|
| HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B27 |
| HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*0101 (DR1), TNFe3, d3, α(−308)1, βn2, a2, b4, HLA-B7 |
| HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*0404 (DR4), TNFe3, d4, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*0101 (DR1), TNFe3, d3, α(−308)2, βn1, a5, b5, HLA-B37 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*1302 (DR13), TNFe3, d3, α(−308)1, βn1, a4, b5, HLA-B7 |
| HLA-DRB1*1601 (DR2), TNFe3, d4, α(−308)1, βn2, a11, b4,HLA-B27 | HLA-DRB1*0401 (DR4), TNFe3, d4, α(−308)1, βn2, a3, b3, HLA-B62 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B44 | HLA-DRB1*0102 (DR1), TNFe3, d3, α(−308)1, βn2, a1, b4, HLA-B44 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B35 | HLA-DRB1*0405 (DR4), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B60 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4, [HLA-B14] | HLA-DRB1*1502 (DR6), TNFe3, d3, α(−308)1, βn1, α2, b4, [HLA-B39] |
| HLA-DRB1*1601(DR2), TNFe3, d4, α(−308)1, βn2, a11, b4, HLA-B35 | HLA-DRB1*0701 (DR7), TNFe3, d3, α(−308)2, βn1, a7, b4,HLA-B13 |
| HLA-DRB1*1601 (DR2), [TNFe3, d3, α(−308)1, βn2, a3, b4, HLA-B51] | HLA-DRB1*0402 (DR4), [TNFe3, d3, α(−308)1, βn2, a6, b4, HLA-B51] |
| HLA-DRB1*1101 (DR11), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B27 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*0701 (DR7), TNFe3, d4, α(−308)1, βn2, a6, b4,HLA-13 | HLA-DRB1*1101 (DR5), TNFe3, d3, α(−308)1, βn2, a6, b5, HLA-B44 |
| HLA-DRB1*0901 (DR9), TNFe3, d3, α(−308)1, βn1, a5, b4, HLA-B7 | HLA-DRB1*0401 (DR4), TNFe3, d3, α(−308)2, βn1, a6, b5, HLA-B44 |
| HLA-DRB1*0401 (DR4), TNFe3, d3, α(−308)1, βn2, a6, b5, HLA-B44 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| TNFe3, d4, α(−308)1, βn2, a7, b4 | TNFe3, d3, α(−308)1, βn2, a7, b4 |
| TNFe3, d3, α(−308)1, βn2, a7, b4 | TNFe3, d4, α(−308)1, βn2, a8, b4 |
| TNFe3, d3, α(−308)1, βn2, a11, b4 | TNFe3, d4, α(−308)1, βn1, a7, b5 |
| TNFe3, d4, α(−308)1, βn2, a10, b4 | TNFe3, d4, α(−308)1, βn1, a5, b5 |
| TNFe3, d3, α(−308)1, βn2, a10, b4 | TNFe3, d4, α(−308)1, βn2, a2, b4 |
| TNFe3, d3, α(−308)1, βn2, a1, b4 | TNFe3, d3, α(−308)1, βn1, a4, b4 |
| First Haplotype . | Second Haplotype . |
|---|---|
| HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B27 |
| HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*0101 (DR1), TNFe3, d3, α(−308)1, βn2, a2, b4, HLA-B7 |
| HLA-DRB1*1501 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*0404 (DR4), TNFe3, d4, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*0101 (DR1), TNFe3, d3, α(−308)2, βn1, a5, b5, HLA-B37 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4,HLA-B7 | HLA-DRB1*1302 (DR13), TNFe3, d3, α(−308)1, βn1, a4, b5, HLA-B7 |
| HLA-DRB1*1601 (DR2), TNFe3, d4, α(−308)1, βn2, a11, b4,HLA-B27 | HLA-DRB1*0401 (DR4), TNFe3, d4, α(−308)1, βn2, a3, b3, HLA-B62 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B44 | HLA-DRB1*0102 (DR1), TNFe3, d3, α(−308)1, βn2, a1, b4, HLA-B44 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B35 | HLA-DRB1*0405 (DR4), TNFe3, d3, α(−308)1, βn2, a11, b4, HLA-B60 |
| HLA-DRB1*1601 (DR2), TNFe3, d3, α(−308)1, βn2, a11, b4, [HLA-B14] | HLA-DRB1*1502 (DR6), TNFe3, d3, α(−308)1, βn1, α2, b4, [HLA-B39] |
| HLA-DRB1*1601(DR2), TNFe3, d4, α(−308)1, βn2, a11, b4, HLA-B35 | HLA-DRB1*0701 (DR7), TNFe3, d3, α(−308)2, βn1, a7, b4,HLA-B13 |
| HLA-DRB1*1601 (DR2), [TNFe3, d3, α(−308)1, βn2, a3, b4, HLA-B51] | HLA-DRB1*0402 (DR4), [TNFe3, d3, α(−308)1, βn2, a6, b4, HLA-B51] |
| HLA-DRB1*1101 (DR11), TNFe3, d3, α(−308)1, βn2, a10, b4, HLA-B27 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| HLA-DRB1*0701 (DR7), TNFe3, d4, α(−308)1, βn2, a6, b4,HLA-13 | HLA-DRB1*1101 (DR5), TNFe3, d3, α(−308)1, βn2, a6, b5, HLA-B44 |
| HLA-DRB1*0901 (DR9), TNFe3, d3, α(−308)1, βn1, a5, b4, HLA-B7 | HLA-DRB1*0401 (DR4), TNFe3, d3, α(−308)2, βn1, a6, b5, HLA-B44 |
| HLA-DRB1*0401 (DR4), TNFe3, d3, α(−308)1, βn2, a6, b5, HLA-B44 | HLA-DRB1*0301 (DR3), TNFe3, d1, α(−308)2, βn1, a2, b3, HLA-B8 |
| TNFe3, d4, α(−308)1, βn2, a7, b4 | TNFe3, d3, α(−308)1, βn2, a7, b4 |
| TNFe3, d3, α(−308)1, βn2, a7, b4 | TNFe3, d4, α(−308)1, βn2, a8, b4 |
| TNFe3, d3, α(−308)1, βn2, a11, b4 | TNFe3, d4, α(−308)1, βn1, a7, b5 |
| TNFe3, d4, α(−308)1, βn2, a10, b4 | TNFe3, d4, α(−308)1, βn1, a5, b5 |
| TNFe3, d3, α(−308)1, βn2, a10, b4 | TNFe3, d4, α(−308)1, βn2, a2, b4 |
| TNFe3, d3, α(−308)1, βn2, a1, b4 | TNFe3, d3, α(−308)1, βn1, a4, b4 |
Haplotypes or generic types with high delta values are in italics. Statistically significant alleles are written in bold. Haplotypes that cannot be assigned are written in brackets.
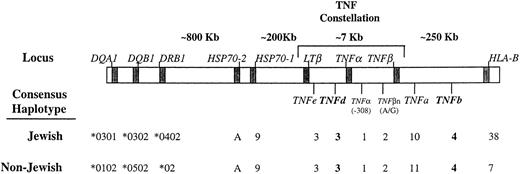
Partial approximate map of the human MHC region with the consensus haplotypes in Jewish and non-Jewish CA patients. TNF alleles with statistically significant differences in frequencies in patients compared with controls are written in bold. The borders for the class-III region are defined by the HLA class II (DRB1 locus) and HLA class I (B locus). The allele assignment for the HSP70-1, -2, and the HLA-DR and HLA-B loci was summarized from our previous publications4 21 (see text for details).
Partial approximate map of the human MHC region with the consensus haplotypes in Jewish and non-Jewish CA patients. TNF alleles with statistically significant differences in frequencies in patients compared with controls are written in bold. The borders for the class-III region are defined by the HLA class II (DRB1 locus) and HLA class I (B locus). The allele assignment for the HSP70-1, -2, and the HLA-DR and HLA-B loci was summarized from our previous publications4 21 (see text for details).
DISCUSSION
Several associations of HLA alleles with drug-induced adverse reactions have been reported in the literature. For example, hydralazine-induced systemic lupus erythematosus (SLE) is associated with HLA-DR4 and the presence of null alleles at the C4 locus.16 Additionally, penicillamine-induced proteinuria and toxicity to chlorpromazine are associated with certain HLA types.29 30
In previous studies we had reported significant association of HLA alleles and haplotypes with CA.4 Additionally, common HSP70 variants were found in CA patients regardless of the ethnic background, suggesting that the genetic susceptibility to develop CA was located in the intermediate portion of the MHC region between the class I and class II loci.22 The associations of HLA alleles and the HSP70 variants were due primarily to the association of two MHC haplotypes: HLA-DRB1*0402, DRB4*0101, DQB1*0302, DQA1*0301, HSP70-2 A, HSP70-1 9.0 in Jewish patients and HLA-DRB1*02, DRB5*02, DQB1*0502, DQA1*0102, HSP70-2 A, HSP70-1 9.0 in non-Jewish patients. However, these haplotypes were not increased significantly in the total population of patients. A possibility that non-HLA alleles in linkage disequilibrium with HLA were involved was raised based on the findings that variants of HSP70 were shared by the two haplotypes associated with CA.4 22
In the present study, we compared the frequencies of polymorphisms of the TNF genes in the group of CA patients with those in a population of 33 white schizophrenic patients who were treated with clozapine without developing agranulocytosis. Susceptibility to CA was associated with a high frequency of the TNF b4 and d3 microsatellite alleles in two different ethnic groups, whereas protection was associated with the microsatellite b5. This is consistent with the hypothesis that susceptibility to this drug-induced reaction is partly due to the linkage disequilibrium of these alleles with HLA-B and DR specificities.4,22-24 The significant association of TNFb4 and d3 in CA patients of Jewish origin could have been predicted as they are part of the extended haplotype HLA-DRB1*0402, DRB4*0101, DQB1*0302, DQA1*0301, HSP70-2 A, HSP70-1 9.0 found in Ashkenazi Jews.15 More informative was the association of TNFb4 and d3 with CA in patients of non-Jewish origin. In 10 of 11 non-Jewish DR2 positive patients and in the remaining 11 patients, there was an association of TNFb4, d3 with CA. This suggests that in non-Jewish patients, the TNF variants are more informative in assigning genetic associations in CA not due to linkage disequilibrium with class I or II alleles. This form of genetic mapping using extended haplotypes or fragments of them (or high delta value haplotypes) has been used successfully in studies of MHC associations with disease.15,21 31
As there is no genetic linkage between the HLA region genes and schizophrenia or resistance to conventional antipsychotic treatment, any alteration of HLA region gene(s) frequencies could not be attributed to these factors. Our findings of an increased frequency of the b4 and d3 TNF microsatellite alleles in the patients suggest that one of these, or a trait determined by another closely linked gene in the intermediate region of the MHC, such as HSP70, is associated with CA. Furthermore, once the disease-causing gene in these ethnic groups is identified, a single direct test can be designed to predict the genetic susceptibility to this disorder. Therefore, the prevention of this adverse drug reaction will allow safer clinical use.
In vitro data on the effects of various cytokines (eg, TNF-α and Interferon-γ) on human hematopoiesis have shown inhibition of the myeloid lineage precursor cells, both slowing their differentiation and inducing their programmed cell death.32-34 In addition, there is in vivo evidence of increased expression of different cytokines in several bone marrow-failure disorders,35 and neutropenia has been reported after the administration of TNFα to healthy individuals.36 There may be a certain mechanism, either immune or toxic, by which clozapine or its metabolites stimulate the production or decrease the clearance of products of the TNF loci in certain genetically susceptible individuals, producing neutropenia or agranulocytosis, depending on the severity of the insult.2 3 However, there are no data at present to support the direct involvement of TNF in the pathogenesis of CA.
It has been established that some HLA haplotypes are associated with a higher production of both TNFα and TNFβ,24,37-39 and it has been suggested that these effects may be mediated by both transcriptional and posttranscriptional regulatory mechanisms.40-42 Additionally, it has been reported that TNFα secretion is influenced either by the TNF microsatellites38,40 or by the HLA-DR generic type.37,38 43 In this regard, studies of neutrophils and of their survival rate in the presence of clozapine or its metabolites could elucidate the role of the TNF constellation polymorphism in TNFα production.
However, it is possible that in individuals carrying risk-associated MHC markers clozapine or its metabolites could induce the expression of HSP70 and/or TNFα and TNFβ, which act as a signal to decrease the proliferative capacity or induce apoptosis in granulocyte precursors or circulating neutrophils.32-34,44,45 Additionally, it may be that certain genetic variants of the different cytokines and their receptors (ie, GM-CSF, IL-2, IL-3) involved in neutrophil development are associated with a different secretion pattern after exposure to clozapine and its metabolites.46
Given the MHC associations found in CA, it would be reasonable to perform HLA phenotyping studies in different idiopathic diseases associated with bone marrow suppression. An illustration of this is the finding that there is a similar genetic background of patients with idiopathic SLE and hydralazine-induced SLE.16 Alternatively, the approach described here could be used by others to find common susceptibility markers on chromosome 6p21 in diseases with different HLA associations.14,15 18
ACKNOWLEDGMENT
We thank Elizabeth Morton and Alison Angel for editorial assistance.
Supported by Grant No. MH-47029 (E.J.Y.) from the National Institute of Mental Health, HL-29583 (C.A.A. and E.J.Y.), and HL-14157 (C.A.A.) from the National Institutes of Health.
Address reprint requests to David Turbay, MD, Department of Pathology, Division of Immunogenetics, Dana-Farber Cancer Institute, 44 Binney St, LHRRB 203, Boston, MA 02115.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal