Abstract
Three novel splice site mutations and two novel missense mutations were identified by molecular analysis of pyruvate kinase (PK) deficiency associated with hereditary nonspherocytic hemolytic anemia. A Nepalese PK variant, PK Kowloon, was found to have a homozygous transversion at the 5′-splice site of the seventh intervening sequence (IVS) of the L-type PK gene (Ivs7[+1]gt → tt). Using a reverse transcription polymerase chain reaction (RT-PCR) assay, we showed that the R-type PK mRNA in the proband's reticulocytes included the seventh IVS between the seventh and eighth exon, introducing a stop codon 3 nucleotides downstream of the mutated site. Consequently, the translational product may lack 44% of the R-PK polypeptide. A transition at the last nucleotide of exon 9 (1269GCG → GCA) was found in a Japanese PK variant, PK ‘Kamata.’ The mutation did not alter the amino acid sequence, but caused skipping of the ninth exonic sequence in the R-PK transcripts. As a result, the affected R-type PK lost 51 amino acid residues (373Met-423Ala del). A transversion at the splice acceptor site of the third IVS (Ivs 3[-2]ag → tg) was identified in PK ‘Aomori.’ The mutation resulted in aberrant splicing at a cryptic splice site within exon 4, causing deletion of two codons in the aberrant R-PK transcript (95 Gly-96 Pro → del). Both PK ‘Kamata’ and PK ‘Aomori’ had a missense mutation on the other allele, 1044AAG → AAT (348Lys → Asn) and 1075CGC → TGC (359Arg → Cys), respectively. Although both 348Lys and 359Arg were located in the sixth loop of A domain (β/α)8 barrel, which has been shown to contain the substrate and cation binding sites, the degree of anemia was much more severe in PK ‘Kamata’ than PK ‘Aomori,’ possibly because the 51 amino acid deletion of PK ‘Kamata’ but the 2 amino-acid deletion of PK ‘Aomori’ may abolish PK catalytic activity.
PYRUVATE KINASE (PK) is a key enzyme of anaerobic glycolysis, which irreversibly catalyzes phosphoenolpyruvate (PEP) to pyruvate. The R (red blood cell)-type PK (R-PK) is the only isoform that is expressed in mature erythrocytes,1 and thus functional alterations of R-PK due to mutations of the L (liver)-type PK (L-PK) gene, which encodes both R- and L-PK2 would cause hemolysis. PK deficiency is the second most common erythroenzymopathy after glucose-6-phosphate dehydrogenase (G6PD) deficiency. This enzymopathy is associated with hereditary nonspherocytic hemolytic anemia, and over 400 cases have been reported to date.3-5 Biochemical characterization of the variant PK has been standardized,6 and most PK variants have been shown to have abnormal kinetics and/or decreased enzymatic stability.7 Recent molecular genetic studies of PK deficiency have demonstrated that most PK variants are caused by heterogeneous missense mutations occurring mainly in the catalytically important domain.8,9 Amino acid changes may alter either hydrophobicity or secondary structure of PK subunits, resulting in decreased affinity for the substrate, PEP,10 as well as impaired responses to allosteric effectors such as fructose-1, 6-diphosphate (FDP).11 X-ray diffraction studies of the cat M1-type isozyme12 showed that the enzyme is a homotetramer and that each subunit is comprised from four domains, namely N, A, B, and C. Data concerning the allosteric transition of PK have been obtained from structural analyses of Escherichia coli PK13 and mutagenesis of Bacillus stearothermophilus PK.14 To elucidate the genotype-phenotype relationship of PK deficiency, we analyzed the enzymatic characteristics and gene mutations in homozygous PK deficiency15 and demonstrated that amino acid substitutions occurred near either the catalytic center or effector binding sites. Decreased affinity for PEP and/or FDP may induce altered allosteric equilibrium, whereas impaired affinity for adenosine diphosphate (ADP) seemed not to be responsible for the deficiency because only one mutation has been reported near the ADP binding site.16 Besides 59 missense mutations including our present data, miscellaneous genetic abnormalities including deletions, insertions, nonsense and splice site mutations, have been reported.9 17 In this report, we describe five novel L-PK gene mutations, including three splice site mutations, and discuss the aberrant R-PK transcripts in the variants' reticulocytes. It is worth describing these mutations, as they have significantly severe phenotypes possibly due to quantitative abnormalities of erythrocyte PK. The mechanisms responsible for the distinct consequences of these splice site mutations will be discussed.
MATERIALS AND METHODS
Materials.Restriction endonucleases were purchased from Takara Shuzo (Kyoto, Japan) and New England Biolabs (Beverly, MA). Taq DNA polymerase (AmpliTaq) was obtained from Perkin Elmer Cetus (Norwalk, CT). Expand reverse transcriptase was purchased from Boehringer (Mannheim, Germany). For the long polymerase chain reaction (PCR) experiment, a commercial kit (LAPCR kit version 2) from Takara Shuzo was used. Oligonucleotides were synthesized with a DNA synthesizer (Applied Biosystems, Model 380A, Foster City, CA). DNA sequence data were obtained using an automated sequencer (Applied Biosystems, Model 373A). The computer software DNASIS (Hitachi Software Engineering, Yokohama, Japan), was used to analyze DNA and amino acid profiles.
Clinical features of PK-deficient subjects.Laboratory data of nonidentical twin Nepalese girls with PK deficiency (PK Kowloon) have been reported elsewhere in detail.18 Intrauterine growth retardation of the twins was noted, and they received exchange blood transfusion during the neonatal period. Hemoglobin levels in the twins at 3 months of age were 1.8 and 2.8 g/100 mL. One of the twins had a recent splenectomy. Histological assessment shows marked hemosiderosis with iron deposition in the hepatocytes. There are no other abnormalities or evidence of cirrhosis.
A Japanese girl (MT, PK ‘Kamata’) received exchange blood transfusion due to severe neonatal jaundice (total bilirubin, 36 mg/100 mL), and was diagnosed as having congenital chronic hemolytic anemia of unknown ethiology. As she required red blood cell (RBC) transfusion once a month, splenectomy was performed at 8 years of age, and since then anemia has been slightly improved with hemoglobin levels of 4-6 to 7-9 g/100 mL. The hematological data at 8 years of age were as follows: RBC count 2.66 × 106/μL, hemoglobin 8.7 g/100 mL, hematocrit 26.1%, and reticulocytes 30%. Serum indirect bilirubin was 6.4 mg/100 mL.
In 1982, an 11-year-old Japanese girl (TF, PK ‘Aomori’) was diagnosed as having congenital hemolytic anemia. The hematological data at 21 years of age were as follows: RBC count 3.36 × 106/μL, hemoglobin 11.1 g/100 mL, hematocrit 32.6%, and reticulocytes 18.8%. Serum indirect bilirubin was 4.8 mg/100 mL.
Enzymatic and biochemical analyses of PK variants.Blood samples were obtained under informed consent from the probands. Because the Nepalese twins had recently been splenectomized, we obtained liver specimens for isozyme and RNA analyses with their parents' informed consent. RBC enzyme activities and glycolytic intermediates were measured by protocols standardized by the International Committee for Standardization in Hematology (ICSH)19 or as described by Minakami et al.20
We separated the reticulocyte-enriched fraction from normochromic RBC by density gradient centrifugation. Heparinized whole blood was passed through a cellulose column to remove white blood cells (WBC) and platelets and then purified RBC were mixed with 19 volumes of density gradient mixture, 35% percoll, 20% urographin, as described.21 Ten-milliliter samples of the suspensions were centrifuged at 35,000g and 4°C for 20 minutes.
RBC PK was partially purified by precipitation with 280 g/L ammonium sulfate, in which both R- and M2-type PK were precipitated. Protein extracts prepared from human liver were homogenized in PK sample buffer containing 10 mmol/L Tris/HCl, pH7.5, 100 mmol/L KCl, 2 mmol/L 2-mercaptoethanol, 10 mmol/L ε-aminocaproic acid, and 10 mmol/L EDTA. Polyclonal antibody against the rat L-PK was kindly provided by Prof T. Noguchi, Fukui Medical School (Fukui, Japan) and used to characterize the isozyme expression of the variant RBC and liver. Aliquots of 100 μg of protein extracts prepared from RBCs and liver were separated on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Proteins were transferred onto a nylon membrane (Hybond N, Amersham, UK) in buffer containing 25 mmol/L Tris, 192 mmol/L glycine, pH 8.3, at 30 V, 4°C, overnight in a mini trans-blot electrophoretic transfer cell (BioRad, Hercules, CA). The membrane was incubated with anti-rat L-PK antibody, followed by chemiluminescence detection using a Western-light Protein Detection Kit (Tropix, Bedford, MA).
Amplification and sequencing of flanking regions of the human L-PK gene.High molecular weight genomic DNA was purified by the standard protocol.22 Each exon of human L-PK gene was amplified by PCR using intronic primers and sequenced as described.15
Structural analysis of R-PK transcripts and genomic sequences.Reticulocyte RNA was purified by the guanidinium isothiocyanate/cesium chloride method.23 Aliquots of 2 μg of total reticulocyte RNA were reverse transcribed in 20-μL reaction mixtures containing 1× Expand reverse transcriptase buffer, 10 mmol/L dithiothreitol, 1 mmol/L deoxyribonucleoside triphosphates (dNTPs), 20 U of ribonuclease inhibitor, 50 pmol of R-1 primer, and 50 U of Expand reverse transcriptase. Aliquots of 1/10 of the cDNA mixture were then subjected to PCR amplification in 50-μL mixtures of 1× LAPCR buffer (included in the kit), 0.4 mmol/L dNTP with 10 pmol each of primers R-2/R-3 and 2.5 U of LATaq DNA polymerase. The reaction mixtures were subjected to 40 cycles of amplification consisting of 94°C for 20 seconds, 60°C for 30 seconds, and 72°C for 210 seconds in GeneAmp PCR System 9600 (Perkin Elmer Cetus). The amplified cDNA spanned 1,819 bp covering the entire coding sequence and parts of the 5′- and 3′-untranslated regions. The amplified product was subsequently subcloned into the pCR II vector using a TA cloning kit (InVitrogen, San Diego, CA), and sequenced using PK gene-specific primers (Table 1) and a Dye termination cycle sequencing kit (Perkin Elmer).
PCR Primers Used in This Study
| To create appropriate restriction sites, the underlined nucleotide were replaced. | |
| R-1 | 5′-CTTAAAGGTGGGGCTTTGGA-3′ |
| R-2 | 5′-CCCAGGCCTACACTGAAAGC-3′ |
| R-3 | 5′-TGTGGGCTGGAGAACGTAGACT-3′ |
| R-4 | 5′-ATCTCCCTAGTGGTCCAGAA-3′ |
| R-5 | 5′-CACCATGATGCCGTCGCTCA-3′ |
| R-6 | 5′-CCAGGAAAACCTTCTCTGCT-3′ |
| R-7 | 5′-AGGACGTCCGAGACCTGCGCTT-3′ |
| R-8 | 5′-TAGCTCCTCAAACAGCTGC-3′ |
| R-9 | 5′-GCGGTGAAGATGCAGCCTGC-3′ |
| R-10 | 5′-GGCGTTCTGAGAAATGGTAATGG-3′ |
| R-11 | 5′-CTCGTGGGAGCCGTGGGAGAAG-3′ |
| R-12 | 5′-AGGTGAGCGACGGCATCATGGT-3′ |
| R-13 | 5′-CATCTTCTGAGCCAGGATCA-3′ |
| R-14 | 5′-CTGTGTGGCACAGACAACAGGCTT-3′ |
| EX-7 | 5′-CCTTTGTGCGGAAAGCCAGC-3′ |
| IVS-7 | 5′-AGGTGATGGGGAATAGCGAC-3′ |
| To create appropriate restriction sites, the underlined nucleotide were replaced. | |
| R-1 | 5′-CTTAAAGGTGGGGCTTTGGA-3′ |
| R-2 | 5′-CCCAGGCCTACACTGAAAGC-3′ |
| R-3 | 5′-TGTGGGCTGGAGAACGTAGACT-3′ |
| R-4 | 5′-ATCTCCCTAGTGGTCCAGAA-3′ |
| R-5 | 5′-CACCATGATGCCGTCGCTCA-3′ |
| R-6 | 5′-CCAGGAAAACCTTCTCTGCT-3′ |
| R-7 | 5′-AGGACGTCCGAGACCTGCGCTT-3′ |
| R-8 | 5′-TAGCTCCTCAAACAGCTGC-3′ |
| R-9 | 5′-GCGGTGAAGATGCAGCCTGC-3′ |
| R-10 | 5′-GGCGTTCTGAGAAATGGTAATGG-3′ |
| R-11 | 5′-CTCGTGGGAGCCGTGGGAGAAG-3′ |
| R-12 | 5′-AGGTGAGCGACGGCATCATGGT-3′ |
| R-13 | 5′-CATCTTCTGAGCCAGGATCA-3′ |
| R-14 | 5′-CTGTGTGGCACAGACAACAGGCTT-3′ |
| EX-7 | 5′-CCTTTGTGCGGAAAGCCAGC-3′ |
| IVS-7 | 5′-AGGTGATGGGGAATAGCGAC-3′ |
To verify the nucleotide changes in the genomic DNA, aliquots of 0.5 μg of genomic DNA were amplified by PCR in 50-μL mixtures of 10 mmol/L Tris/HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2 , 0.001% gelatin, 0.2 mmol/L dNTP with 25 pmol of each primer listed in Table 2, and 1.25 U of AmpliTaq DNA polymerase. The reaction mixtures were subjected to 30 cycles of amplification consisting of 94°C for 20 seconds, 60°C for 20 seconds, and 72°C for 20 seconds in a GeneAmp PCR System 9600. The amplified DNA was digested with the restriction endonucleases listed in Table 2 to verify the nucleotide changes found in the cDNA.
Strategy for Detection of L-PK Gene Mutations
| . | Primers . | PCR Products (bp) . | Restriction Enzyme . | Normal Allele . | Mutant Allele . |
|---|---|---|---|---|---|
| Ivs7[+1]gt→tt | R-6/R-7 | 351 | Hph I | 178 + 88 + 58 + 27 | 205 + 88 + 58 |
| 1269GCG→GCA | R-8/R-9 | 170 | Pst I | 115 + 55 | 95 + 55 + 20 |
| Ivs 3[−2]ag→tg | R-10/R-11 | 179 | Bfa I | 95 + 84 | 179 |
| 1044AAG→AAT | R-12/R-13 | 83 | Hph I | 60 + 14 + 9 | 69 + 14 |
| 1075CGC→TGC | R-12/R-14 | 134 | Hha I | 94 + 40 | 134 |
| . | Primers . | PCR Products (bp) . | Restriction Enzyme . | Normal Allele . | Mutant Allele . |
|---|---|---|---|---|---|
| Ivs7[+1]gt→tt | R-6/R-7 | 351 | Hph I | 178 + 88 + 58 + 27 | 205 + 88 + 58 |
| 1269GCG→GCA | R-8/R-9 | 170 | Pst I | 115 + 55 | 95 + 55 + 20 |
| Ivs 3[−2]ag→tg | R-10/R-11 | 179 | Bfa I | 95 + 84 | 179 |
| 1044AAG→AAT | R-12/R-13 | 83 | Hph I | 60 + 14 + 9 | 69 + 14 |
| 1075CGC→TGC | R-12/R-14 | 134 | Hha I | 94 + 40 | 134 |
Southern blot hybridization of the RT-PCR products.Aliquots of reticulocyte cDNA were amplified in the same reaction solution used for genomic DNA amplification described above. Primers R-4 and R-5 flanking exon 7 were used. The PCR products were separated on a 1.5% agarose gel and transferred onto a nylon membrane. Hybridization was performed in 5× SSC (1× SSC is 0.15 mol/L NaCl, 0.015 mol/L sodium citrate, pH 7.0), 1% blocking reagent (supplied with DIG Nucleic Acid Detection Kit, Boehringer), 0.1% N-lauroylsarkosine, 0.02% SDS at 60°C. The final washing conditions were 65°C in 0.1× SSC-0.1% SDS. Oligonucleotide probes, EX-7 and IVS-7 were labeled at their 3′ ends with digoxigenin (DIG oligonucleotide 3′-end labeling kit) and used as probes. Positive bands were detected by chemiluminescence (DIG nucleic acid detection kit; Boehringer Mannheim Biochemica, Mannheim, Germany).
RESULTS
Homozygous 5′-splice site mutation of IVS 7 identified in PK Kowloon.RBC enzyme analysis of the PK Kowloon family showed almost normal PK activity in both twins (twin 1, 13.5; twin 2, 11.1 IU/g Hb, normal range, 12.0 to 15.6), whereas PK activity of the mother was decreased (9.03 IU/g Hb). It seemed most likely that there was contamination by normal RBC in the probands' blood, because the twins remained transfusion-dependent. To exclude PK activity derived from normal RBCs, we separated reticulocytes and young RBCs by density gradient centrifugation using percoll/urographin21 and measured PK activity. PK activity of the fractionated RBCs of twin 1 was decreased to below the normal range, to about 75% of the normal mean value, 10.4 IU/g Hb.
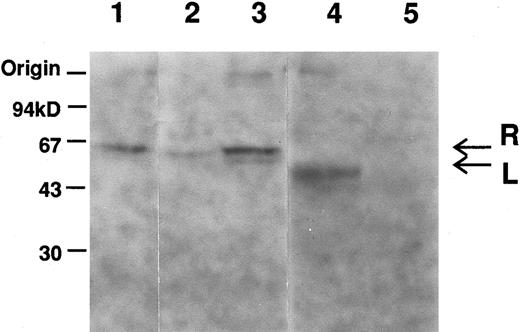
Immunoblot analysis showed a decrease in amount of the R-PK subunit in the reticulocyte fraction (Fig 1, lane 2), but not in unfractionated samples (Fig 1, lane 3). The L-PK subunit was not detected in the liver extract (lane 5).
Immunoblot analysis of erythrocytes and liver extracts with anti-L–PK. Labeled bands, which migrated at positions corresponding to about 62 (arrow designated as R) and 60 (arrow designated as L) kD, were R-and L-PK, respectively. Lane 1, normal erythrocytes; lane 2, low density fraction of erythrocytes of PK Kowloon; lane 3, unfractionated erythrocytes of PK Kowloon; lane 4, normal liver; lane 5, liver of PK Kowloon.
Immunoblot analysis of erythrocytes and liver extracts with anti-L–PK. Labeled bands, which migrated at positions corresponding to about 62 (arrow designated as R) and 60 (arrow designated as L) kD, were R-and L-PK, respectively. Lane 1, normal erythrocytes; lane 2, low density fraction of erythrocytes of PK Kowloon; lane 3, unfractionated erythrocytes of PK Kowloon; lane 4, normal liver; lane 5, liver of PK Kowloon.
Genomic DNA sequencing showed that the variant had a transversion at +1 position of the 5′-splice site of intron 7 (Ivs 7[+1] gt → tt). As the mutation destroyed an Hph I restriction site, the genomic DNAs of the family members, as well as a normal control, were amplified by PCR using primers R-6/R-7, and digested with Hph I. Both twins were homozygous for this mutation, while the parents were heterozygotes (data not shown). To elucidate the effect of the mutation on RNA splicing, we amplified the 1.7-kb R-PK cDNA of PK Kowloon. Agarose gel electrophoresis demonstrated two RT-PCR products of different sizes. One of the RT-PCR products had the normal R-PK cDNA sequence, while the other cDNA with a slightly larger molecular weight showed an insertion of 96 nucleotides, which was identical to the seventh IVS between exons 7 and 8. Southern blot hybridization of the RT-PCR products flanking exon 7 of the L-PK gene showed an additional larger cDNA corresponding to IVS 7 in PK Kowloon, as well as a 336-bp cDNA corresponding to the normally spliced transcript (Fig 2, lane 1). The oligonucleotide complementary to the IVS 7 (Fig 2, lanes 3 and 4) hybridized with the 432-bp cDNA in PK Kowloon (lane 3), but not in the control (lane 4). These observation led us to conclude that the 5′-splice site mutation caused aberrant splicing of the primary transcript of the L-PK gene, resulting in insertion of the IVS 7 in the mature transcript. Although both the R-PK subunits and transcripts were detected in reticulocytes of the patient, it is likely that most of the molecules were derived from contaminating normal RBC and reticulocytes due to recurrent blood transfusion, because L-PK expression was hardly observed in the patient's liver as shown by immunoblotting (Fig 1). Translation of the aberrant mRNA terminates prematurely in the IVS 7 (AGttga). The expected length of the truncated R-PK subunit was 322 amino acid residues, meaning that the polypeptide lost 44% of the R-PK. We were unable to detect the short R-PK subunit immunologically.
Southern blot hybridization of RT-PCR products. Oligo-dT primed reticulocyte RNA was used for PCR with primers R-4 and R-5 flanking exon 7 of the R-PK cDNA. Normal R-PK cDNA is 336 bp long, which includes 25, 271, and 40 bp cDNA sequences of exon 6, 7, and 8. If the 96 bases of the seventh intervening sequence are retained, the aberrant cDNA becomes 432 bp long. Oligonucleotide EX-7 hybridized with exon 7 (lane 1, PK Kowloon, lane 2; control), and IVS-7 hybridized with intron 7 (lane 3, PK Kowloon, lane 4; control).
Southern blot hybridization of RT-PCR products. Oligo-dT primed reticulocyte RNA was used for PCR with primers R-4 and R-5 flanking exon 7 of the R-PK cDNA. Normal R-PK cDNA is 336 bp long, which includes 25, 271, and 40 bp cDNA sequences of exon 6, 7, and 8. If the 96 bases of the seventh intervening sequence are retained, the aberrant cDNA becomes 432 bp long. Oligonucleotide EX-7 hybridized with exon 7 (lane 1, PK Kowloon, lane 2; control), and IVS-7 hybridized with intron 7 (lane 3, PK Kowloon, lane 4; control).
1269A mutation identified in PK ‘Kamata’ is responsible for the skipping of exon 9 sequence.RBC enzyme analysis showed that PK activity of the proband was decreased to 44% of the normal mean value (6.0 IU/g Hb). The PK activity of the mother was also below the normal range (11.6 IU/g Hb). Glycolytic intermediates upstream of PK were accumulated in the proband: 2,3-diphosphoglycerate (2,3-DPG), 12320 (normal range, 3,438 to 5,218); 3-phosphoglycerate (3PG), 185.9 (25.6 to 55.2); 2-phosphoglycerate (2PG), 41.9 (3.0 to 11.0); PEP 152.8 (8.9 to 19.7)(nmol/mL RBC).
The reticulocyte R-PK cDNA of PK ‘Kamata’ amplified with R-2/R-3 primers showed double bands of about 1.7 and 1.6 kb. Sequencing of a short cDNA indicated that the R-PK cDNA lacked exon 9. The skipping of exon 9 caused a deletion of 51 amino acids (373Met-423Ala del). These amino acid residues included residues essential for the function of the enzyme, and thus the variant PK may have no activity. To explore the molecular abnormality accounting for the exon skipping, we analyzed cDNA and genomic DNA sequences of PK ‘Kamata’ and found that the proband was a compound heterozygote of 1269GCG → GCA (423Ala → Ala)/1044AAG → AAT (348Lys → Asn, K348N). These mutations were verified by PCR-restriction fragment length polymorphism (RFLP) analysis using the primers and restriction endonucleases listed in Table 2. The 1269 G → A mutation does not alter the amino acid sequence, but is responsible for aberrant splicing at the 5′-splice site of IVS 9. 348 Lys is evolutionarily conserved from chicken through humans and is located in the sixth alpha helix of the A domain (Aα6) of the PK subunit.12 Recently Mattevi et al13 elucidated the three-dimensional structure of the allosteric type PK of E coli and demonstrated that 292Arg on the Aα6, which corresponded to 337Arg of the human R-PK, participated in substrate binding in the allosterically active state. K348N may cause decreased affinity for PEP due to conformational changes of the active site.
Novel splice acceptor site mutation identified in PK ‘Aomori.’RBC enzyme assay of the proband showed that PK activity was decreased to 49% of the normal mean value (6.7 IU/g Hb). Glycolytic intermediates upstream of PK were accumulated: 2,3-DPG, 7560; 3PG, 121.5; 2PG, 14.4; PEP 44.2 (nmol/mL RBC). Sequencing of the R-PK cDNA and L-PK gene showed that the variant was a compound heterozygote of two novel point mutations; Ivs 3 [-2]ag → tg and 1075 CGC → TGC (359 Arg → Cys, R359C). The splice acceptor site mutation induced cryptic splice site utilization in the 5′-end sequence of exon 4 (ag → tgGGCCAG). Consequently, the R-PK transcript lacked the first six nucleotides of exon 4, resulting in deletion of two amino acids in the R-PK (95 Gly-96 Pro → del). R359C may cause decreased affinty for PEP, because the substituted 359Arg was located in the same alpha helix (Aα6)12 as 348 Lys.
DISCUSSION
Approximately 15% of point mutations accounting for human genetic diseases cause aberrant mRNA splicing.24 In RBC enzymopathies, splicing defects are known to be the common abnormalities in phosphofructokinase (PFK) deficiency.25 Aberrant splicing may reduce or diminish mRNA levels, as it can cause exon skipping, nucleotide insertion/deletion or premature translational termination. In PK deficiency, seven splice site mutations have been identified to date including those in the present cases.11,15,26,27 40 However, none have been analyzed at the cDNA level. In this study, we described three splicing defects of the human L-PK gene associated with PK deficiency, and by analyzing reticulocyte PK mRNA structure, we confirmed that each mutation resulted in a distinct molecular abnormality.
A Nepalese PK variant, PK Kowloon, is one of the most severe PK deficiencies so far reported. The probands were true homozygotes of the 5′ splice site mutation at IVS 7. Splice donor site mutations at +1 position have been reported to cause either skipping of preceded exons or utilization of cryptic splice sites.28,29 In PK Kowloon, however, only the transcripts that retained seventh IVS were detected. We searched for potential splice sites using statistical scoring according to the theory of Shapiro and Senepathy.30 Using the nucleotide sequence data around 5′-splice sites of 97 human genes,31 the 5′-splice site of the IVS 7 scored as 90.6, and the adjacent 5′ and 3′ GT sequences counted as 57.0 or 45.0, respectively. Even with gt → tt mutation, the score was 73.4, which may still be sufficient for efficient splicing at the mutated site and, therefore, cryptic sites were not used. The skipping of exon 7 would not occur, possibly because the length of exon 7, 271 bp, is much longer than that of IVS 7, 96 bp. Thus, the splice donor site mutation may cause retention of IVS 7.
Two PK isozymes, the L- and M2-type, expressed in normal liver,32 and those isozymes were encoded by distinct structural genes.33 Although PK Kowloon had virtually no L-PK in liver, the M2-PK activity had been detected by polyacrylamide gel electrophoresis (data not shown). Biochemical data of the probands showed the increments of serum transaminases, alkaline phosphatase, as well as hyperbilirubinemia. We believed that these hepatic abnormalities were likely due to hemosiderosis, but not to the PK deficiency in the liver.
Several examples have been reported of nucleotide substitutions occurring at the last base of exons being a cause of aberrant splicing.34,35 Ohshima and Gotoh31 have shown statistically that 80% of the last nucleotides of 97 donor sites of human genes were guanine. Although both G → A and G → C27 at 1,269 of the L-PK gene caused small decreases in the splicing score, the skipping of exon 9 may have occurred due to the relatively short (94 bp) intronic length.
Mutations of the invariant ag (−2) at 3′-splice sites have been reported to be responsible for usage of cryptic sites36 or exon skipping.37 The splice acceptor site mutation at IVS 3 of PK ‘Aomori’ caused cryptic utilization of the exonic sequence within exon 4, resulting in a two-amino acid deletion near the amino-terminal region of the affected PK subunit. Because amino acid substitutions have not been identified in this region, with only one exception,16 we speculate that the variant PK with the two amino-acid deletion might show catalytic activity; a similar situation was reported in a molecular study of G6PD deficiency. An 8-amino acid deletion was identified in a Japanese G6PD variant, G6PD Nara.38 The variant enzyme had residual activity even with the deletion, and Rawland39 recently showed that the region where the amino acid deletion occurred was not essential for G6PD enzymatic activity by analyzing the tertiary structure of the Leuconostoc homolog. This speculation was supported by the observation that accumulation of glycolytic intermediates in erythrocytes, as well as the degree of anemia of PK ‘Aomori,’ seemed to be much milder than those in PK ‘Kamata,’ although the missense mutations at the other alleles were supposed to have similar effects on the catalytic activity.
The present study has unveiled the molecular lesions of severe PK deficiency with marked structural alterations of the affected PK subunit. The consequences of the mutations were demonstrated by analyzing reticulocyte mRNA structure. Reticulocyte RNA seems a useful source of experimental material for analyzing molecular defects of severe hemolytic anemia due to PK deficiency.
NOTE ADDED IN PROOF
Bianchi et al40 reported a splice site mutation recently.
ACKNOWLEDGMENT
We thank Y. Okamura, A. Sakuma, M. Watanabe, and J. Oka for their technical assistance. We are also indebted to Dr J.C. Wilson for managing the patient and providing the blood sample.
Supported, in part, by a Scientific Research Grant from the Ministry of Education, Science, Sports, and Culture, by the Mitsui Life Social Welfare Foundation, Tokyo, and by a Research Grant for Specific Diseases from the Ministry of Health and Welfare, Tokyo, Japan.
Address reprint requests to Hitoshi Kanno, MD, PhD, Okinaka Memorial Institute for Medical Research, 2-2-2 Toranomon, Minato-ku, Tokyo 105, Japan.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal