Abstract
Epstein-Barr virus (EBV) is a human lymphotropic virus whose main targets have traditionally been described as B lymphocytes and epithelial cells. Here we report the isolation and characterization of largely monoclonal transformed human T-cell lines infected by EBV. The transformed T cells expressed CD2, CD3, and either CD4 or CD8 surface molecules and more generally displayed the phenotype of naive T cells with a complete and clonal rearrangement of the T-cell receptor. None of the cell lines expressed B cells, natural killer, or myeloid antigens or had immunoglobulins genes rearrangement. They grew in the absence of growth factor; however, they all secreted interleukin-2 after mitogenic activation. Polymerase chain reaction (PCR) analysis showed the presence of EBV DNA in all these cell lines. Moreover, Southern blot analysis of one of these cell lines shows the presence of circular episomic EBV DNA, and by Northern blot or reverse transcriptase-PCR analysis, only the expression of Epstein-Barr nuclear antigen-1 (EBNA-1) and latent membrane protein-1 (LMP-1) genes was detected. Finally, the complete transformed phenotype of this T-cell line was shown by its injection into nude or recombination activating gene 2 (RAG2)-deficient mice that led to the formation of solid tumors.
THE EPSTEIN-BARR virus (EBV) is a human herpes virus that causes infectious mononucleosis and is believed to be etiologically associated with Burkitt's lymphoma,1 nasopharyngeal carcinoma,2 and Hodgkin's disease.3 It has recently been reported that EBV infection is correlated not only to B-cell disorders, but also to nasal T-cell lymphoma,4,5 angioimmunoblastic lymphadenopathy (AILD)-like T-cell lymphoma,6,7 and peripheral T-cell lymphoma in immunocompetent hosts8,9 in both EBV-endemic and nonendemic areas. However, despite the growing number of descriptions of T-cell lymphoma infected by EBV, very few EBV-infected T cells have been isolated and characterized in vitro so far. There are two reports on the establishment and the characterization of EBV-infected T cells isolated from patients,10,11 showing the relation between EBV and T-cell growth in vivo. However, a possible secondary infection by EBV of already proliferating T cells cannot be excluded. In addition, two different groups12,13 using a recombinant EBV expressing a selectable maker have shown that the MT2 T-cell line was infectable by EBV, however, as the MT2 cell line was already transformed by the human T-cell leukemia virus-1 (HTLV-1) virus, they could not analyze the transformation potency of EBV on T cells. The only report that illustrates the potential oncogenic properties of EBV on T cells described a T-cell line obtained after transfection of EBV genome into cord blood lymphocytes.14 However, the oncogenic mechanism was not analyzed in that report, and the mechanism does not seem to be highly reproducible.
The cellular receptor for the EBV (CD21, CR2),15 a 145-kD integral transmembrane single-chain glycoprotein that functions also as a receptor for the surface-bound C3dg and C3d fragments of human C316 was thought to be expressed only on mature B lymphocytes and follicular dendritic cells. However, recent studies have shown that a molecule identical or very similar to CD21 is expressed on a population of human T cells and thymocytes.17,18 In addition, a fraction of T lymphocytes and some T-cell lines have been shown to express functional EBV receptors, and experimental EBV infection of these cells has been documented.10,19 20
EBV is also able to establish lymphoblastoid cell lines (LCLs) after infection in vitro of peripheral blood mononuclear cells.21 It has been described that EBV-LCL were able to secrete T-cell restricted lymphokines such as interleukin (IL)-2.22 However, whether this IL-2 secretion was produced by the EBV-infected B cells themselves or by contaminating T cells was not established in that study. We describe here the isolation, from EBV-LCLs obtained from different donors, of several EBV-infected transformed T-cell lines.
MATERIALS AND METHODS
Cells and culture conditions.EBV-LCLs were obtained as previously described.23 EBV-LCLs, as well as the different T-cell lines, were cultured in RPMI containing 20% fetal calf serum, human transferrin (10 μg/mL; GIBCO-BRL, Gaithersburg, MD), and bovine insulin (10 μg/mL; GIBCO-BRL).
Non-T cells, non-B cells, and T cells were purified by using a cocktail of antibodies specific for CD2, CD3, CD4, CD8, CD19, CD20, and CD56, respectively. T cells were purified by negative depletion with CD19, CD20, CD14, HLA-DR, CD56 antibodies. Positively labeled cells were removed with dynabeads as specified by the manufacturer (Dynal, Oslo, Norway). In non-T, non-B cells, less than 5% of contaminated B or T cells were detected and in purified T cells, the purity was greater than 98% as analyzed with a CD3 monoclonal antibody (MoAb).
Electroporation experiments.Electroporation experiments were performed according to a protocol previously established for Jurkat cells. A total of 107 cells in 250 μL opti-MEM (GIBCO-BRL) were incubated with 10 μg plasmid in a cuvette on ice for 15 minutes. Cells were then electroporated at 250 V, 1200 μF, for 60 milliseconds. Immediately after electroporation, cells were transferred in complete culture medium. This electroporation protocol, which is far too stringent for EBV-LCL B cells, induced more than 99% cell death after 1 day of culture. Cells were plated in 24-well plates, and the medium was replaced every 2 days. After 4 to 5 weeks, few living cells were detected in some wells. They finally grew out into permanently transformed cell lines about 2 months after the beginning of the experiment.
Determination of lymphokine production.Cells were cultured at 1 × 106 cells/mL with medium alone or with CD3 MoAb cross-linked on plates at 1 μg/mL in 0.1 mol/L Tris pH 9.5 buffer plus tetradecanoyphorbol-13-acetate (TPA; 1 ng/mL) or with cross-linked CD3 MoAb and CD28 MoAb (1 μg/mL) as previously described.24 Supernatants were collected after 24 hours, and the presence of IL-2 was quantified by immunoenzymetric assay (Genzyme, Cambridge, MA) according to the manufacturer's instructions.
Immunofluorescence and immunochemistry analysis.For detection of cell surface antigens, 105 cells were labeled with phycoerythrin (PE)- or fluorescein isothiocyanate (FITC)-conjugated MoAbs (all antibodies came from Becton Dickinson, Sunnyvalle, CA, except for CD21 from Caltag Laboratories, South San Francisco, CA). Cells were incubated for 30 minutes with the appropriate antibody at 4°C in phosphate-buffered saline (PBS) with 0.1% bovine serum albumin (BSA), 0,02 mmol/L NaN3. After three washes, the labeled cell samples were analyzed on a FACScan (Becton Dickinson).
Immunofluorescence and immunochemistry experiments were performed on adhesion slides (Bio-Rad, Ivry sur Seine, France) using a standard ABC technique. Briefly, 10 μL of a cell suspension at 106 cells/mL was layered on each spot of the slide. Cells were fixed for 15 minutes in PBS, 3% formaldehyde. All further incubations were performed in Hanks' Balanced Salt Solution (HBSS), 0.01 mol/L HEPES, and 0.1% Saponin. Slides were blocked by a blocking step with horse serum (10% in HBSS-saponin) followed by a blocking step with an Avidin D solution for 15 minutes followed by incubation with the biotin solution according to the manufacturer's instructions (Vector, Burlingame, CA). Cells were then incubated for 45 minutes with the primary antibody specific for Epstein-Barr nuclear antigen-1 (EBNA-1; 6F9, mouse IgG2; Biogenesis, Poole, UK), EBNA-2 (PE2, mouse IgG1; Biogenesis), latent membrane protein-1 (LMP-1; mix, mouse IgG1; Biogenesis), or a mix of mouse IgG1 and IgG2 antibodies (Becton Dickinson, San Jose, CA), respectively. Mouse MoAbs were labeled for 30 minutes with biotinylated horse antimouse immunoglobulins (Vector). After washing steps, biotin was revealed either with an ABC peroxidase enzyme system (Vector Elite Standard ABC kit; Vector) for immunochemistry analysis or with an ABC phycoerythrin system (Vectastain ABC-phycoerythrin; Vector) for immunofluorescence analysis. CD3 expression on T cells was analyzed with an anti-CD3 MoAb (Becton Dickinson, France) revealed with a fluoresceinated antimouse F(ab′)2 fragments. Fluorescence was analyzed on a confocal microscope (Aristoplan-CLSM Leica, France).
Western blot analysis.Western blot analysis was performed according to techniques already published.25 Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% tricine gels; Novex, San Diego, CA), proteins were transferred onto membranes (Immobilon-P; Millipore, St Quentin en Yuelines, France) by electroblotting. Membranes were blocked with 5% dry nonfat milk in PBS and then incubated with the primary antibody directed against EBNA-1 (6F9, mouse IgG2; Biogenesis) for 18 hours. Following washing, membranes were incubated with a peroxidase-conjugated goat antimouse serum (Biosource International, Camarillo, CA) revealed with the ECL kit (Amersham, France). Blots were autoradiographed for 2 minutes.
Reverse transcription.Total RNA was purified using RNAzol according to the manufacturer's instructions (Bioprobe, Montreuil Sous Bois, France). Before reverse transcription, RNA samples were treated with DNAse I (GIBCO-BRL) to remove any cellular or viral DNA contaminant. A total of 2 μg of RNA was treated with 2 U of DNAse I RNAse-free for 15 minutes at room temperature. The DNAse was then inactivated by the addition of 1 μL of 25 mmol/L EDTA and samples were heated for 10 minutes at 65°C. Reverse transcription was performed as described.26 Briefly, 2 μg of total RNA in 9 μL were mixed with 4 μL buffer (5× RT buffer: 250 mmol/L Tris-HCl pH 8.3, 300 mmol/L KCl, 15 mmol/L MgCl2), 1 μL primer (0.5 μg oligo dT), 4 μL deoxynucleotide triphosphate (dNTP; 10 mmol/L), 0.1 μL dithiothreitol (1 mol/L), 0.1 μL RNAsin (40 U/μL), 0.1 μL BSA (1 μg), 1 μL Escherichia coli rRNA (4 μg), 0.8 μL Moloney murine leukemia virus reverse transcriptase (MMLV-RT; 80 U). This mixture was centrifuged and incubated at 42°C for 1 hour. Samples were heat denatured at 95°C for 3 minutes and after rapid cooling, 50 U in 2 μL of MMLV-RT was added and a second cycle of reverse transcription performed.
PCR analysis.For PCR amplification, 1 μL of the reverse transcriptase reaction was added to a total reaction volume of 25 μL of the PCR mix (1× PCR buffer, 200 μmol/L of each dNTP, 0.5 μmol/L of each primer, 2 mmol/L MgCl2, and 2.5 U/100 μL Taq polymerase). After an initial denaturation step at 94°C for 2 minutes, 35 cycles of PCR were performed (1 minute at 55°C, 1 minute at 72°C, and 1 minute at 94°C). A 20-μL aliquot was run on a 2% agarose gel containing ethidium bromide.
To analyze EBV-specific sequences, the PCR was performed using primer pairs specific for EBNA-1 5′: AGCGATAGAGCAGGGCCCCGCAGAT, 3′: CAAAACCTCAGCAAATATATGAGTT amplifying a 430-bp fragment or specific for EBNA-2 5′: CTATCTTGCGTTACATGGGGGACA 3′: GGGAGTGGTGGGGGCACCCCC amplifying a 177-bp fragment or specific for LMP-1 5′: CACGACCTTGAGAGGGGCCCA 3′: GCCAGATGGTGGCACCAAGTC amplifying a 406-bp fragment on cDNA and a 562-bp on DNA. Analysis of viral DNA sequence in infected cells was performed by directly lysing the cell according to the method already described.27 For the preparation of cell lysates containing genomic DNA, cells were diluted in PBS at 1 × 107 cells/mL. One microliter (containing 10,000) was added to 9 μL of distilled water into a 96-well plate and then 20 μL of PCR buffer was added (50 mmol/L KCl, 10 mmol/L Tris, HCl, pH 8.3, 2.5 mmol/L MgCl2, 1 mg/mL gelatin, 0.05 mg/mL proteinase K, 20 mmol/L dithiothreitol, and 0.7 μmol/L SDS). Complete lysis was observed under phase contrast microscopy. Lysates were then transferred to a 500-μL tube, vortexed for 30 seconds, incubated for 1 hour at 37°C, incubated for 10 minutes at 86°C, and stored at −20°C.
Twenty microliters of cell lysate containing the genomic DNA was introduced in a 500-μL tube and 80 μL of PCR buffer was added. After an initial denaturation step at 94°C for 10 minutes, 35 cycles of PCR were performed (30 seconds at 60°C, 30 seconds at 72°C, and 30 seconds at 94°C).
Analysis of immunoglobulin heavy chain gene rearrangement by PCR.Consensus primers used to amplify the IgH gene were: FR3A: ACACGGC(C,T)(C,G)TGTATTACTGT for the 3′ end of the V region and LJH: TGAGGAGACGGTGAC and VLJH: GTGACCAGGGT(A,C,G,T)CCTTGGCCCCAG for the 3′ end of the J region. For amplification of the Ig gene, a seminested PCR was performed: a first round of 20 cycles (94°C, 30 seconds; 58°C, 30 seconds; 72°C, 30 seconds) with primers LJH and FR3A, followed by a 1:2500 dilution, and then a second round of 30 cycles with primers VLJH and FR3A. The PCR was performed on 0.1 μg of DNA purified by phenol chloroform extraction using 0.4 U Taq polymerase (Amplitaq; Roche Molecular Systems, Branchburg, NJ) in the presence of anti-Taq antibody (Clontech, Palo Alto, CA). Each PCR round was preceded by a 5-minute denaturation at 94°C and was followed by a final extension of 7 minutes at 72°C. Fragments were electrophoresed through 2% agarose gels in Tris borate acetate (TBE) buffer.
As a control for DNA integrity, we used specific primers inside an exon of the Cμ region of immunoglobulins and the Cβ region of the T-cell receptor (TCR). The Cβ-specific primers that amplified a 250-bp fragments were GTGTTTGAGCCATCAGAAGCAGAG and AGAAATCCTTTCTCTTGACCATGGCCA. The Cμ-specific primers that amplified a 350-bp fragment were: ATCACTTTCTCCTGGAAATA and TGGGACGAAGACGCTCACTTTGGGA.
Southern blotting.Southern blot experiments were performed according to standard techniques.28 All DNA was digested with EcoRI. The LMP probe used represents the entire 1,100 bp DNA sequence of the LMP1 gene and was cloned by PCR with the TA cloning kit (Invitrogen, San Diego, CA). The human Cβ1 cDNA probe, a 496-bp cDNA specific for the Cβ1 region, was obtained by PCR and cloned using the TA cloning kit.
Northern blotting.Northern blot experiments were performed as previously described.24 The EBNA-1 and the LMP-1–specific probes were obtained by PCR with the primers described above and using EBV-LCL cDNA as a template.
RESULTS
Isolation of EBV-infected transformed T-cell lines.In an attempt to transfect and select long-term established EBV-transformed B-cell lines (EBV-LCLs) with a plasmid expressing different human immunodeficiency virus (HIV) proteins and the hygromycinR gene under the thymidin kinase promoter,29 we surprisingly isolated some T-cell lines among the transfected B cells. To control that these T cells were not transformed by the expression of any transfected HIV-protein and to test the potential role of the selective drug, further electroporation experiments were performed on EBV-LCLs using a pSV2-neoR vector, which encodes only for the neoR gene. After 4 to 5 weeks of culture in a medium containing G418 (10 μg/mL), we isolated living cells and some of them finally grew out as T-cell lines after 2 months (Table 1). The majority of the other cell lines displayed a B-cell phenotype (not shown). This result was not due to a particular donor, as electroporation of three EBV-LCLs from three different donors repeatedly allowed the isolation of T cells (described in Table 1). Electroporation in the absence of DNA or culture of cells electroporated with the plasmid in the absence of selecting drug led to the isolation of only transformed B cells. These results suggest that the enrichment for T cells obtained after this transfection procedure, which was originally set up to transfect Jurkat cells, may be due to the preferential transfection of the plasmid into T cells that are then selected with the drug. As a control, lipofection of the EBV-LCLs using the same plasmid with (or without selective drugs) did not led to isolation of T cells (not shown).
Cell Surface Phenotype of Different EBV-Infected Transformed T-Cell Lines
| . | CD2 . | CD3 . | TCR . | CD4 . | CD8 . | CD28 . | CD25 . | CD45RA . | CD45RO . | CD56 . | HLA-DR . | CD44 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EBV 41 | 97 | 78 | 73 | 3 | 68 | 100 | 0 | 85 | 10 | 0 | 0 | 5 |
| EBV 32 | 93 | 94 | 69 | 7 | 97 | 100 | 0 | 87 | 5 | 3 | 0 | 3 |
| T1B5 | 99 | 92 | 94 | 0 | 87 | 100 | 6 | 89 | 7 | 0 | 0 | 6 |
| TFS | 90 | 97 | 89 | 87 | 0 | 100 | 4 | 92 | 2 | 0 | 0 | 14 |
| E1V | 85 | 81 | 97 | 47 | 0 | 100 | 0 | 87 | 6 | 0 | 0 | 10 |
| 2C8 | 99 | 95 | 79 | 94 | 0 | 100 | 5 | 93 | 7 | 2 | 0 | 6 |
| 2G4 | 95 | 97 | 91 | 94 | 0 | 100 | 12 | 96 | 9 | 3 | 0 | 5 |
| 2G8 | 96 | 94 | 42 | 67 | 4 | 100 | 0 | 99 | 7 | 4 | 0 | 0 |
| NC5 | 98 | 99 | 96 | 3 | 98 | 100 | 2 | 97 | 10 | 3 | 0 | 3 |
| TC | 100 | 98 | 95 | 9 | 94 | 100 | 6 | 89 | 9 | 0 | 0 | 9 |
| . | CD2 . | CD3 . | TCR . | CD4 . | CD8 . | CD28 . | CD25 . | CD45RA . | CD45RO . | CD56 . | HLA-DR . | CD44 . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EBV 41 | 97 | 78 | 73 | 3 | 68 | 100 | 0 | 85 | 10 | 0 | 0 | 5 |
| EBV 32 | 93 | 94 | 69 | 7 | 97 | 100 | 0 | 87 | 5 | 3 | 0 | 3 |
| T1B5 | 99 | 92 | 94 | 0 | 87 | 100 | 6 | 89 | 7 | 0 | 0 | 6 |
| TFS | 90 | 97 | 89 | 87 | 0 | 100 | 4 | 92 | 2 | 0 | 0 | 14 |
| E1V | 85 | 81 | 97 | 47 | 0 | 100 | 0 | 87 | 6 | 0 | 0 | 10 |
| 2C8 | 99 | 95 | 79 | 94 | 0 | 100 | 5 | 93 | 7 | 2 | 0 | 6 |
| 2G4 | 95 | 97 | 91 | 94 | 0 | 100 | 12 | 96 | 9 | 3 | 0 | 5 |
| 2G8 | 96 | 94 | 42 | 67 | 4 | 100 | 0 | 99 | 7 | 4 | 0 | 0 |
| NC5 | 98 | 99 | 96 | 3 | 98 | 100 | 2 | 97 | 10 | 3 | 0 | 3 |
| TC | 100 | 98 | 95 | 9 | 94 | 100 | 6 | 89 | 9 | 0 | 0 | 9 |
Results are expressed as the percentage of positive cells. Cells were stained with saturating amounts of FITC- or PE-conjugated MoAbs (Becton Dickinson) and analyzed by using a cytofluometer (Epics; Becton Dickinson). EBV 41 and EBV 32 were isolated from donor 1; T1B5, TFS, and E1V from donor 2; 2C8, 2G4, and 2G8 and NC5 from donor 3. TC cells were isolated from a solid tumor in a RAG2-deficient after subcutaneous injection of NC5 cells.
To confirm the presence of contaminating T cells in different EBV-LCLs that have been cultured for more than 1 year, we performed RT-PCR experiments using two different sets of primers specific for the Cα and the Cβ chains of the TCR, respectively.30 We detected a signal for both the Cα and the Cβ transcripts in all seven EBV-LCLs tested (not shown), demonstrating that the presence of T cells in EBV-LCLs is not a rare event.
We then tried to use T-cell specific MoAbs to purify these T cells. We also tried to clone these T cells by using end point dilution techniques. However, their low frequency into EBV-LCLs, as well as the finding that these T cells died when they were too diluted, resulted in failure to isolate them by these techniques.
Finally, infection by EBV of resting or activated purified peripheral blood T cells from the same donors failed to promote the transformation of T cells.
Immortalized T cells have a phenotype of naive T cells.All T-cell lines were characterized as homogeneous round cells growing in suspension. They were cultured for more than 2 years in the absence of any growth factor showing their immortalized phenotype. As mentioned above, however, these cells proliferated very poorly (except for the NC5 cell line that was used in most of the experiments) and required culturing at a high cell density. All cell lines are exclusively composed of T cells expressing the TCR, CD2, CD3, CD28, and either CD4 or CD8 surface molecules (Table 1), but no B cell (CD19, CD20) NK (CD16 or CD56), or monocyte (CD14), specific surface antigens. Surprisingly, for transformed T-cell lines, they showed a naive phenotype, as they were CD45RA+, CD45RO−. In addition, they did not express HLA-DR, CD69, CD44, and CD25 (Table 1).
Analysis of the clonality and the rearrangement of the TCR.The clonality and the rearrangement of the TCR genes was evidenced by performing a Southern blot on two of these immortalized T-cell lines named NC5 (Fig 1A) and TC (derived from NC5 after in vivo injection into RAG-/-mice), using a Cβ1 specific probe. Purified non-T non-B peripheral mononuclear cells, the monocytic cell line U937, or an EBV-LCL cell line (lanes 1, 4, and 5, respectively) displayed the germline pattern with two germline bands of 12 (Cβ1) and 4.2 kb (Cβ2), respectively, following digestion with EcoRI. The specific additional band with a length of approximately 10 kb observed in both Jurkat cells, TC, and NC5 cells indicates a complete clonal rearrangement of the TCR genes and furthermore shows the mature differentiated character of these immortalized T cells.
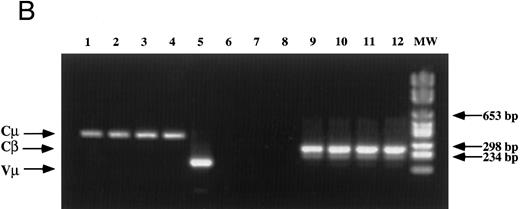
Analysis of TCR and immunoglobulin gene rearrangements in EBV-transformed T cells. (A) EcoRI digested NC5 (lane 2) and TC (lane 3) DNA and control DNA from purified non-T, non-B human peripheral mononuclear cells (lane 1), monocytic cell line U937 (lane 4), EBV-LCL (lane 5), purified peripheral blood T cells (lane 7), and T-cell lymphoma Jurkat (lane 8) were analyzed with a 496-bp human Cβ1 cDNA probe. The sizes of the Cβ-germline restriction fragments obtained from non-T cells were 12 and 4.2 kb for Cβ1 and Cβ2, respectively (large arrows). The additional rearranged band (small arrows) of approximately 10 kb detected in EBV-transformed T cells (lanes 2 and 3) and in Jurkat cells (lane 8) showed the clonality of these cell populations, whereas in polyclonal purified T cells from peripheral blood, the mixture of all possible rearrangements is shown by a smear (lane 7). The molecular weight markers (MW II; Boehringer Mannheim) were loaded in lane 6. (B) The rearrangement of immunoglobulin heavy chains was analyzed by using primers specific for the VJ region of immunoglobulins amplifying a 105-bp fragment of DNA from an EBV-LCL (lane 5), the EBV-transformed T cell NC5 (lane 6), the EBV-transformed T cell TC (lane 7), or Jurkat cells (lane 8). To control for DNA integrity primers amplifying a 350-bp fragment inside an exon of the Cμ heavy chain region (lanes 1 to 4) or primers amplifying a 250-bp inside an exon of the Cβ chain of the TCR (lanes 8 to 12) were used on the DNA from an EBV-LCL (lanes 1 and 9), the EBV-transformed T cell NC5 (lanes 2 and 10), the EBV-transformed T cell TC (lanes 3 and 11), or Jurkat cells (lanes 4 and 12).
Analysis of TCR and immunoglobulin gene rearrangements in EBV-transformed T cells. (A) EcoRI digested NC5 (lane 2) and TC (lane 3) DNA and control DNA from purified non-T, non-B human peripheral mononuclear cells (lane 1), monocytic cell line U937 (lane 4), EBV-LCL (lane 5), purified peripheral blood T cells (lane 7), and T-cell lymphoma Jurkat (lane 8) were analyzed with a 496-bp human Cβ1 cDNA probe. The sizes of the Cβ-germline restriction fragments obtained from non-T cells were 12 and 4.2 kb for Cβ1 and Cβ2, respectively (large arrows). The additional rearranged band (small arrows) of approximately 10 kb detected in EBV-transformed T cells (lanes 2 and 3) and in Jurkat cells (lane 8) showed the clonality of these cell populations, whereas in polyclonal purified T cells from peripheral blood, the mixture of all possible rearrangements is shown by a smear (lane 7). The molecular weight markers (MW II; Boehringer Mannheim) were loaded in lane 6. (B) The rearrangement of immunoglobulin heavy chains was analyzed by using primers specific for the VJ region of immunoglobulins amplifying a 105-bp fragment of DNA from an EBV-LCL (lane 5), the EBV-transformed T cell NC5 (lane 6), the EBV-transformed T cell TC (lane 7), or Jurkat cells (lane 8). To control for DNA integrity primers amplifying a 350-bp fragment inside an exon of the Cμ heavy chain region (lanes 1 to 4) or primers amplifying a 250-bp inside an exon of the Cβ chain of the TCR (lanes 8 to 12) were used on the DNA from an EBV-LCL (lanes 1 and 9), the EBV-transformed T cell NC5 (lanes 2 and 10), the EBV-transformed T cell TC (lanes 3 and 11), or Jurkat cells (lanes 4 and 12).
The absence of any immunoglobulin heavy chain gene rearrangement was analyzed by PCR as previously described31 (Fig 1B). This result showed the absence of any immunoglobulin genes rearrangement in the transformed T cells, ruling out the possibilities of either a fusion between T cells and immortalized B cells during electroporation, as well as the selection of immortalized B cells that would have subsequently rearranged their TCR genes and expressed T-cell specific proteins on their cell surface. The absence of cell fusion was also ruled out by a karyotype analysis that showed a normal number a chromosomes (C. Montpellier et al, manuscript submitted).
Transformed T cells are infected by EBV.The association of EBV with these transformed T cells was analyzed by different techniques. The presence of viral DNA was detected by PCR in all T-cell lines using different pairs of primers specific for EBNA-1 (Fig 2A), EBNA-2, LMP-1, or for the BamH1 W region and the Dhet region (not shown).
Detection of EBV DNA in EBV-transformed T cells. (A) EBV DNA was detected by PCR analysis on DNA extracted from seven different EBV-transformed T-cell lines (EBV 41, T1B5, TFS, 2C8, 2G4, 2G8, NC5, respectively, lanes 1 to 7), a T-cell lymphoma Jurkat (lane 8), or two EBV-LCLs (lanes 9 and 10), or a control without DNA (lane 11) using primer pairs specific for LMP-1 amplifying a 562-bp fragment. (B) The presence of EBV DNA in EBV-transformed T cells (NC5 and TC) was confirmed by Southern blot hybridization on 10 μg of EcoRI digested genomic DNA (lanes 1 to 4) or on BamHI digested Hirt extract from 2 × 106 cells (lanes 6 to 9) with a LMP-1 specific probe. Lanes 4 and 9, EBV LCL; lanes 3 and 8, Jurkat cells; lanes 2 and 7, NC5 cells; lanes 1 and 6, TC cells; lanes 5 and 10, molecular weight markers II (Boehringer Mannheim, France).
Detection of EBV DNA in EBV-transformed T cells. (A) EBV DNA was detected by PCR analysis on DNA extracted from seven different EBV-transformed T-cell lines (EBV 41, T1B5, TFS, 2C8, 2G4, 2G8, NC5, respectively, lanes 1 to 7), a T-cell lymphoma Jurkat (lane 8), or two EBV-LCLs (lanes 9 and 10), or a control without DNA (lane 11) using primer pairs specific for LMP-1 amplifying a 562-bp fragment. (B) The presence of EBV DNA in EBV-transformed T cells (NC5 and TC) was confirmed by Southern blot hybridization on 10 μg of EcoRI digested genomic DNA (lanes 1 to 4) or on BamHI digested Hirt extract from 2 × 106 cells (lanes 6 to 9) with a LMP-1 specific probe. Lanes 4 and 9, EBV LCL; lanes 3 and 8, Jurkat cells; lanes 2 and 7, NC5 cells; lanes 1 and 6, TC cells; lanes 5 and 10, molecular weight markers II (Boehringer Mannheim, France).
The infection of the transformed NC5 T cells by EBV was confirmed by Southern blot analysis with a LMP-1 specific probe (Fig 2B). Analysis of Hirt extracts32 showed a band of episomic DNA in both the EBV-LCL (Fig 2B, lane 9) and the EBV-infected immortalized T-cell lines (Fig 2B, lanes 6 and 7). Moreover, the size of the BamHI fragment (greater than 10 kb) detected with the LMP-1 probe encompassing the 31 terminal end of the genome is compatible with a circular configuration of this episomal DNA. However, the analysis of the EcoRI digested genomic DNA showed a specific band only in the EBV-LCL sample (Fig 2B, lane 4) and not in the EBV-infected transformed T-cell samples (Fig 2B, lanes 1 and 2), suggesting that no viral integration occurred in these T cells. As expected under the conditions used here, the probe did not hybridize EBV-negative Jurkat cells (Fig 2B, lanes 3 and 8) and normal T lymphocytes (not shown).
EBV-infected transformed T cells expressed EBNA-1 and LMP-1 gene products.The expression of EBV-specific mRNAs was investigated by Northern blotting experiments (Fig 3A and B) and RT-PCR (Fig 3C). In both cases, only transcripts coding for the EBNA-1 and LMP-1 genes of EBV were detected in EBV-immortalized T-cell lines (Fig 3), whereas in EBV-transformed B-cell lines, we detected transcripts coding for EBNA-1, LMP-1, EBNA-2 genes (Fig 3), but also for EBNA-3a, b, c genes (not shown).
Detection of EBV transcripts in EBV-transformed T-cell lines. (A) and (B) Northern blot analysis on 10 μg of total RNA isolated from the EBV-transformed T cell NC5 (lane 1), the EBV-transformed T cell TC (lane 2), a T-cell lymphoma Jurkat (lane 3), a EBV-LCL (lane 4), or peripheral blood mononuclear cells (B, lane 5) was performed using a 470-bp cDNA fragment specific for EBNA-1 (A) or a 406-bp cDNA fragment specific for LMP-1 (B). An equal amount of RNA was loaded in each lane as controlled by the intensity of the 28S and 18S bands after ethidium bromide labeling. (C) The detection of the EBV transcripts was performed by RT-PCR using the EBNA-1 primers amplifying a 470-bp fragment, the EBNA-2 primers amplifying a 177-bp fragment, and the LMP-1 primers amplifying a 406-bp fragment, described above on, respectively, an EBV-LCL (lanes 1 to 3), an EBV-transformed T cell NC5 (lanes 4 to 6,) an EBV-transformed T cell TC (lanes 7 to 9), or a T-cell lymphoma Jurkat (lanes 10 to 12). As a control for DNA contamination, RNA samples that were processed identically, but in which the reverse transcriptase had been omitted, were amplified by PCR, no positive bands were detected.
Detection of EBV transcripts in EBV-transformed T-cell lines. (A) and (B) Northern blot analysis on 10 μg of total RNA isolated from the EBV-transformed T cell NC5 (lane 1), the EBV-transformed T cell TC (lane 2), a T-cell lymphoma Jurkat (lane 3), a EBV-LCL (lane 4), or peripheral blood mononuclear cells (B, lane 5) was performed using a 470-bp cDNA fragment specific for EBNA-1 (A) or a 406-bp cDNA fragment specific for LMP-1 (B). An equal amount of RNA was loaded in each lane as controlled by the intensity of the 28S and 18S bands after ethidium bromide labeling. (C) The detection of the EBV transcripts was performed by RT-PCR using the EBNA-1 primers amplifying a 470-bp fragment, the EBNA-2 primers amplifying a 177-bp fragment, and the LMP-1 primers amplifying a 406-bp fragment, described above on, respectively, an EBV-LCL (lanes 1 to 3), an EBV-transformed T cell NC5 (lanes 4 to 6,) an EBV-transformed T cell TC (lanes 7 to 9), or a T-cell lymphoma Jurkat (lanes 10 to 12). As a control for DNA contamination, RNA samples that were processed identically, but in which the reverse transcriptase had been omitted, were amplified by PCR, no positive bands were detected.
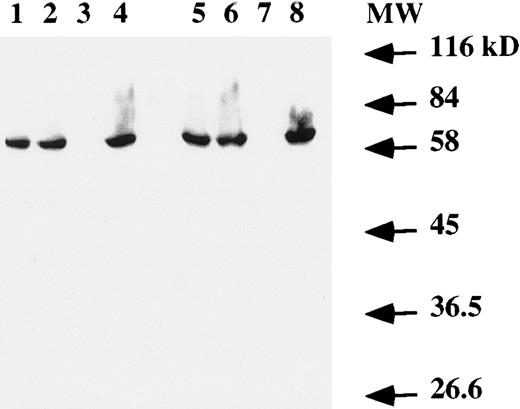
Western blot analysis on total cell extracts showed the presence of the EBNA-1 protein (Fig 4) in both T and B cells infected by EBV. No specific band was detected in the Jurkat cell line (Fig 4, lanes 3 and 7).
Western blot analysis of EBNA-1 expression. Total protein extract from 106 (lanes 1 to 4) or 2 × 106 (lanes 5 to 8) cells were electrophoresed on a SDS-PAGE gel. After transfer to a membrane, EBNA-1 protein was revealed with an anti-EBNA–1 monoclonal antibody. Lanes 1 and 5, EBV-transformed T-cell line NC5; lanes 2 and 6, EBV-transformed T-cell line TC; lanes 3 and 7, T-cell lymphoma Jurkat; and lanes 4 and 8, EBV-LCL. The numbers on the right denote the standard molecular weights in kD.
Western blot analysis of EBNA-1 expression. Total protein extract from 106 (lanes 1 to 4) or 2 × 106 (lanes 5 to 8) cells were electrophoresed on a SDS-PAGE gel. After transfer to a membrane, EBNA-1 protein was revealed with an anti-EBNA–1 monoclonal antibody. Lanes 1 and 5, EBV-transformed T-cell line NC5; lanes 2 and 6, EBV-transformed T-cell line TC; lanes 3 and 7, T-cell lymphoma Jurkat; and lanes 4 and 8, EBV-LCL. The numbers on the right denote the standard molecular weights in kD.
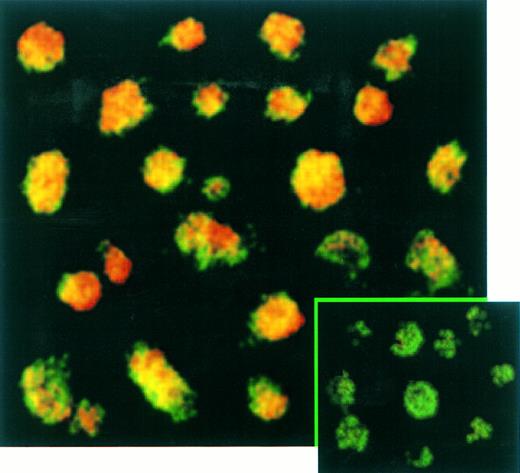
The percentage of T cells productively infected by EBV was measured by immunofluorescence (Fig 5) and immunochemistry (not shown) assays. These experiments confirmed the absence of EBNA-2 expression in EBV-infected transformed T cells (not shown) and showed that LMP-1 (Fig 5) and EBNA-1 (not shown) were expressed in virtually 100% of these T cells labeled with a fluorescein-anti–CD3 monoclonal antibody. No positive signal by both immunochemistry or immunofluorescence assays was observed on Jurkat cells labeled with anti-EBNA–1, anti-EBNA–2, or anti-LMP–1 MoAbs as expected (not shown).
Confocal analysis of double immunofluorescence assay on EBV-transformed T cells. EBV transformed T cells (NC5) were labeled with the anti-LMP–1 MoAb revealed by a phycoerythrin-ABC complex and by an anti-CD3 MoAb revealed by a fluorescein-goat antimouse immunoglobulin F(ab)′2. Cells labeled with the IgG1 control isotype and the CD3 MoAbs are shown in the bottom left corner as a control. Cells were analyzed with a confocal microscope. The green color represents the fluorescein-labeled CD3 MoAb, the red color represents the phycoerythrin-labeled anti-LMP–1 MoAb, and the yellow color represents the mixture of the two colors as detected by the confocal microscope.
Confocal analysis of double immunofluorescence assay on EBV-transformed T cells. EBV transformed T cells (NC5) were labeled with the anti-LMP–1 MoAb revealed by a phycoerythrin-ABC complex and by an anti-CD3 MoAb revealed by a fluorescein-goat antimouse immunoglobulin F(ab)′2. Cells labeled with the IgG1 control isotype and the CD3 MoAbs are shown in the bottom left corner as a control. Cells were analyzed with a confocal microscope. The green color represents the fluorescein-labeled CD3 MoAb, the red color represents the phycoerythrin-labeled anti-LMP–1 MoAb, and the yellow color represents the mixture of the two colors as detected by the confocal microscope.
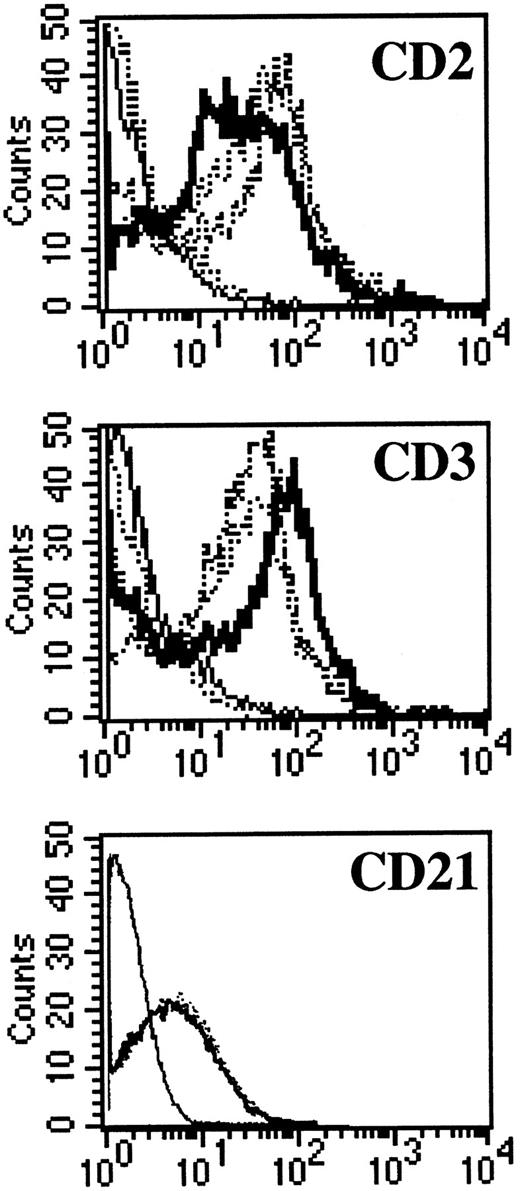
NC5 cells have a malignant phenotype.We analyzed the ability of the EBV-infected transformed T-cell line, NC5, to induce tumors in immunodeficient mice. Injection of NC5 cells into nude mice or RAG2-deficient mice repeatedly led to the formation of solid tumors in 80% to 90% of the mice injected (not shown). We isolated the cells of one of these tumors (TC cells) and compared them with the cells initially injected (NC5). For all the different parameters analyzed, phenotype (Table 1, Fig 6), TCR (Fig 1), EBV infection, and gene expression (Figs 2 and 3), no difference was detected between the cells isolated from the tumor and the cell line transformed in vitro. Moreover, none of the tumor cells reacted with H-2Kd–specific antibodies eliminating the possibility of contaminating mouse T cells. Finally, the expression of CD21, a likely candidate for EBV receptor on T cells, was analyzed on both cell lines. Figure 6 shows that CD21 expression on these EBV-transformed T-cell lines was equivalent to the expression on a EBV-LCL, indicating that infection of the T cells was obtained through that receptor.
CD3, CD2, and CD21 expression on NC5 and TC cell lines. One EBV B-cell line (negative dotted line), Jurkatt T-cells (thick black line), and the EBV-transformed T-cell lines NC5 and TC (positives dotted lines) were analyzed for expression of the CD2 and CD3 T-cell–specific antigens. The isotype control is shown as a straight black line. In addition, the EBV B-cell line and the two EBV-transformed T cells, NC5 and TC, were analyzed for the expression of CD21. The different histograms for the three different lines were completely indistinguishable.
CD3, CD2, and CD21 expression on NC5 and TC cell lines. One EBV B-cell line (negative dotted line), Jurkatt T-cells (thick black line), and the EBV-transformed T-cell lines NC5 and TC (positives dotted lines) were analyzed for expression of the CD2 and CD3 T-cell–specific antigens. The isotype control is shown as a straight black line. In addition, the EBV B-cell line and the two EBV-transformed T cells, NC5 and TC, were analyzed for the expression of CD21. The different histograms for the three different lines were completely indistinguishable.
The transformed T cells secrete IL-2 after activation.We investigated the properties of the different immortalized T-cell lines by measuring their cytokine profiles after polyclonal activation (Table 2). Although these T-cell lines did not secrete detectable amounts of IL-2, stimulation with different mitogenes induced the secretion of IL-2 (Table 2) without inducing a further enhancement in their proliferation rate (not shown). This induction of IL-2 secretion after mobilization of the TCR indicates that these transformed T cells might be useful as new tools for analyzing T-cell activation mechanisms.
Activation Induces IL-2 Secretion in Different EBV-Transformed T-Cell Lines
| T-Cell Lines . | Medium . | CD3 MoAb + TPA . | CD3 + CD28 MoAbs . |
|---|---|---|---|
| 2C8 | <0.01 | 7.5 ± 1.3 | NT |
| TFS | <0.01 | 5.4 ± 2.2 | NT |
| NC5 | <0.01 | 6.2 ± 0.9 | 3.4 ± 0.4 |
| TC | <0.01 | 5.8 ± 1.4 | 2.2 ± 1.3 |
| T-Cell Lines . | Medium . | CD3 MoAb + TPA . | CD3 + CD28 MoAbs . |
|---|---|---|---|
| 2C8 | <0.01 | 7.5 ± 1.3 | NT |
| TFS | <0.01 | 5.4 ± 2.2 | NT |
| NC5 | <0.01 | 6.2 ± 0.9 | 3.4 ± 0.4 |
| TC | <0.01 | 5.8 ± 1.4 | 2.2 ± 1.3 |
Cells were cultivated at 1 × 106 cells/mL with medium alone or with plastic bound CD3 MoAb (1 μg/mL) plus TPA (1 ng/mL) or with plastic bound CD3 and CD28 MoAb (1 μg/mL). Supernatants were collected after 24 hours and the presence of IL-2 was quantified by immunoenzymetric assay. Results represent the mean of 3 different analysis performed in triplicates and are expressed in μg/mL.
Abbreviation: NT, not tested.
DISCUSSION
In this report, we describe the establishment of transformed T-cell lines infected by EBV isolated from peripheral blood mononuclear cells infected in vitro. Immunofluorescence analysis (Table 1, Figs 5 and 6) have shown that all the different cell lines isolated are composed exclusively of T cells. Moreover, Southern blot analysis using a TCR Cβ-specific probe (Fig 1) has proven for at least one of them their oligoclonality. All of the T cells in the different cell lines are infected by EBV as detected by PCR (Fig 2) and at least in one of them (NC5), the expression of the EBV proteins EBNA-1 and LMP-1 is documented by immunoflurorescence (Fig 5) and immunochemistry (not shown). Their transformed and malignant phenotype is shown by their proliferation in the absence of any growth factors and by the ability of NC5 to develop as a tumor in immunodeficient mice. Finally, the expression of the CD21 molecule on the NC5 cell line (Fig 6) suggest that this molecule is a likely route for infection, although this has not yet been directly proved.
At this stage, it is not possible to affirm that the transformation state of the T cells is the result of the presence and expression of the virus. Indeed, no direct infection and transformation of purified T cells by EBV was obtained in vitro so far. However, the isolation of EBV-infected transformed T cells was established in cells from three different donors (Table 1) after infection with EBV in vitro, and none of these cells was positive for HTLV-1 DNA tested by PCR,33 or for the T lymphotropic Saimiri Herpes tested by Southern blot experiments using a Saimiri Herpes virus C-region specific probe.34 Moreover, there are striking homologies between the T cells we have isolated and the other chronically EBV-infected T cells described so far. No evidence for integrated EBV DNA or linear EBV DNA was observed in the NC5 cell line, a finding that is in accordance with the reports on either EBV-infected T cells isolated from patients,10 or in vitro infection of T-cell lines,12 suggesting that in contrast to thymocytes infected in vitro,35 the cell lines described here are at a latent phase of the infection.
In addition, the different EBV-infected transformed T cells expressed only EBNA-1 and LMP-1 EBV genes, whereas EBV-infected B cells from the same donors expressed nine different latent proteins including three LMP and six EBNA genes. EBNA-1 is believed to be essential for viral replication in an episomic state of EBV,36 whereas EBNA-2 and LMP-1, which are expressed in B-LCLs, are thought to be responsible for the establishment and maintenance of the immortal stage.37 In recent studies, Fujiwara and Ono12 and Yoshiyama et al,13 using recombinant EBV to infect the HTLV-I immortalized MT2 T-cell line, have shown that EBV-infected MT2 cells expressed only the EBNA-1 and LMP-1 genes. In peripheral T-cell lymphoma,7,38 as well as in nasopharyngeal carcinoma39 and Hodgkin's disease,40,41 RT-PCR experiments showed the presence of EBNA-1 and LMP-1 transcripts, in the absence of EBNA-2 transcripts.38 Finally, a more recent study analyzing EBV-infected IL-2–dependent T-cell lines isolated from severe chronic active EBV-infected patients, also shows the restricted protein expression in EBV-infected T cells.10 All of these studies are in accordance with our results on EBV-infected transformed T cells obtained in vitro. Together these data favor the hypothesis of a unique pattern of viral expression in T cells restricted to EBNA-1 and LMP-1 as an obligatory program, but also as a contribution to the EBV-linked T-cell transforming process.
Of the viral proteins that are essential for transformation of B cells, LMP-1 is of particular interest, as it resembles a classical oncogene in its ability to induce growth transformation of rodent fibroblasts,42 to morphologically transform human keratinocytes in vitro,43 to inhibit human epithelial cell differentiation,44 or to induce DNA synthesis and expression of activation markers in B45 or T lymphocytes.46 Recently, LMP-1 has been shown to engage signaling proteins for the tumor necrosis factor (TNF)-R family.47 Thus, through constitutive interaction with TNF receptor-associated factors, LMP-1 may amplify or usurp physiologic signal transduction and promote cell transformation. Another hypothesis for the role of LMP-1 in the transformation process is its ability to induce upregulation of the bcl-2 oncogene. The expression of Bcl-2 promotes survival of B-cell precursors and of memory and immunoglobulin-secreting B cells,48 and it also blocks programmed cell death in B cells or T cells.49 Further characterization of the biological and chemical effect of LMP-1 on human T cells will probably be relevant to understand its effect in EBV-induced T-cell transformation.
The most important difference between EBV-immortalized B cells and the EBV-infected transformed T cells we isolated is the malignant phenotype of the T cells as shown by their tumorigenicity in immunodeficient mice. In EBV-infected B cells, it has been described that expression of IL-6, after stable transfection of an IL-6–expressing vector, led to an altered pattern of growth and to a malignant phenotype, confirmed by tumorigenicity in nude mice.50 Similarly, overexpression of IL-2 in murine T-cell clones induces a malignant phenotype characterized by the formation of solid tumors in immunodeficient mice after injection of IL-2–overexpressing T cells.51 However, EBV-transformed T-cell lines did not secrete detectable amounts of IL-2 in the absence of activation (Table 2) or IL-4, IL-5, IL-6, IL-10, IL-13, TNF-α, or interferon-γ (not shown), ruling out a possible role for one of these cytokines in the malignant phenotype of EBV-transformed T cells.
In conclusion, although more work is required to really ascertain the critical role that EBV may play in promoting tumor onset as opposed to simple opportunistic presence, these new EBV-infected transformed T cells can be very useful tools in vitro to further analyze the EBV T-cell interactions and EBV-induced oncogenicity in T cells. Also, these new T-cell lines could be useful in analyzing the T-cell activation process or as targets for viral infection and viral production like HIV or hepatitis B virus.
ACKNOWLEDGMENT
The authors thank Josiane Henno for her technical assistance and Kevin Bacon for reviewing this manuscript.
Address reprint requests to Hervé Groux, PhD, DNAX Research Institute of Immunology and Cellular Biology, 901 California Ave, Palo Alto, CA 94304.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal