Abstract
Seven secondary leukemia patients were treated for solid tumors or malignant lymphoma with anticancer drugs or radiation. We studied bone marrow samples from these patients by fluorescence in situ hybridization (FISH). Of the seven patients, three had increased signals for the ABL oncogene (9q34) on interphase nuclei and at metaphase. One of the three patients also had four signals for the CD3 (MLL) region (11q23). Whole painting probes revealed that these chromosomal regions were translocated onto structurally abnormal chromosomes, resulting in partial tri-, tetra- or penta-somy of these regions. We called this type of translocation “segmental jumping translocation (SJT).” SJT of the ABL oncogene was not detected in samples from 15 patients with de novo acute myelocytic leukemia (AML), 12 with myelodysplastic syndrome (MDS), or 20 with chronic myelocytic leukemia (CML) at the chronic phase. Furthermore, monosomy 7 was also found in the patients with the gene amplification. These results indicate that SJT of ABL and/or CD3 (MLL) genes is associated with the leukemogenesis of secondary leukemia. The SJT may be one mechanism of gene amplification.
ONCOGENES ARE OFTEN amplified in advanced solid tumors. The amplified DNA can be observed either as an extended chromosome, called the homogeneously staining region (HSR) or as extra chromosomal elements, referred to as double minute (dmin) chromosomes.1 However, the incidence of HSR and dmin in leukemia is extremely rare. Among samples from 300 patients with acute myelocytic leukemia (AML) studied in our laboratory, only three had dmin chromosomes accounting for approximately 1% of AML.2 An increased risk of therapy-related AML and myelodysplastic syndrome (MDS) has been reported in the patients treated with alkylating agents for diseases such as non-Hodgkin's lymphoma, multiple myeloma, and solid tumors.3-6 The therapy-related leukemias morphologically and cytogenetically differ from de novo acute leukemias.7,8 Monosomy 7 and structural abnormalities of chromosomes 5 and 7 are common in secondary leukemias.9 10
The association of oncogene activation in secondary leukemia has not been investigated in detail. Here, we studied seven patients with secondary leukemia, by means of conventional chromosome analysis and by fluorescence in situ hybridization (FISH). Four of the seven had complex chromosome aberrations with multiple marker chromosomes, in which chromosomal segments with the ABL oncogene were partially duplicated in three, including one with duplication of the ABL oncogene and CD3 regions. These chromosomal segments translocated onto structurally abnormal chromosomes or unidentifiable marker chromosomes. We named this phenomenon “segmental jumping translocation (SJT).” This is a hitherto undiscovered cytogenetic mechanism for gene amplification and it is seen quite frequently in secondary leukemias, especially in those with highly complex karyotypes.
MATERIALS AND METHODS
Patients.Seven patients were diagnosed with secondary leukemia after extensive chemotherapy and/or radiation therapy between 1990 and 1995. One patient had been treated for non-Hodgkin's lymphoma and 6 for solid tumors including 3 uterus and 1 each for breast, laryngeal, and skin cancers. Two patients (patients no. 1 and 5) had been exposed to the atomic bomb in Hiroshima, with the estimated radiation doses at 5.18 and 0.16 Gy, respectively.
Cytogenetic and FISH analysis.Cytogenetic studies were performed on bone marrow cells using 24-hour cultures and G-banding. Karyotypes were determined according to the International System for Human Cytogenetic Nomenclature (ISCN).11 The same samples that were G-banded were also analyzed by FISH. Air-dried smears were prepared by dropping the cell suspensions onto glass slides. The slides were dried at room temperatures for 1 to 2 days, then hybridized with a biotinylated ABL probe (Oncor Science, Gaithersburg, MD) and with biotinylated D9S7 cosmid probe (Japan Gene Bank, Tokyo, Japan). A digoxigenin-labeled M-BCR probe (Oncor Science) was the hybridization control. Whole chromosome painting probes of chromosome 9 and 11 were obtained from Vysis (Naperville, IL). A CD3 YAC probe containing the CD3 and MLL genes, and which is located on the breakpoint of the 11q23 translocation in acute leukemia, was provided by Dr M. Seto (Aichi Cancer Center, Nagoya, Japan). Monosomy 7 was detected with the α-satellite, D7Z1 probe on the centromeric region of chromosome 7 (Oncor Science). FISH was essentially performed as described.12
Hybridized signals were scored in about 100 to 500 interphase nuclei for each patient under a fluorescence microscope using appropriate absorption and excitation filters (Olympus, Tokyo, Japan). Nuclei with unclear morphology or ambiguous signals were excluded from scoring. The percentage of nuclei with more than two signals was evaluated as gene amplification. Cut-off levels of ABL and D7Z1 probes were estimated from seven patients without hematologic diseases. The values were 8.1% (mean ± 3SD) and 12.2% (mean ± 3SD) in ABL and D7Z1 probes, respectively.
Clinical, Cytogenetic, and FISH Results of Seven Secondary Leukemia Patients
| Patient No. . | Sex/Age . | Primary Tumor . | Secondary Leukemia . | Therapy* . | Duration (yrs)=d . | Survival Time (mo) . | Monosomy 7 (%) . | Total Cells . | No. of ABL Gene . | ABL Gene Amplification (%) . | CD3 Amplification . | Karyotype . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | . | . | . |
| 1 | F 71 | Uterus | AML M2 | ;gg-Rays | 6.3 | 5 | 89.3 | 183 | 0 | 60 | 87 | 29 | 6 | 1 | 0 | 67.2 | ;ms | 41,XX,;ms5,;ms7,;ms15,;ms20,;ms21,;ms22,;ms22,;pl2r;ob10;cb/41,XX,;ms5.;ms7,;ms11,;ms16,;ms19,;ms20,;ms21,;plmar1, ;plmar2;ob9;cb.nuc ish 9q34(ABLx2-4) |
| 40Gy | ||||||||||||||||||
| CP | ||||||||||||||||||
| 2 | F 67 | Uterus | AML M4 | ;gg-Rays | 5.5 | 8 | 27.4 | 219 | 0 | 210 | 2 | 4 | 3 | 0 | 0 | 4.1 | ;ms | 46,XX;ob20;cb.nuc ish 9q34(ABLx2) |
| 3 | F 70 | Uterus | AML M2 | ;gg-Rays | 9.7 | 17 | 4.6 | 221 | 0 | 213 | 0 | 8 | 0 | 0 | 0 | 3.6 | ;ms | 46,XX;ob22;cb.nuc ish 9q34(ABLx2) |
| 4=D | F 63 | Breast | AML M1 | 5-FU | 7.3 | 2.3 | 5.0 | 272 | 0 | 66 | 83 | 27 | 80 | 14 | 2 | 75.7 | ;pl | 44,XX,dup(1)(q25q44),del(4)(q25q28),del(5) (q13q31),i(11)(q10),der(12)t(11;12)(q23;p12), del(13)(q12q14),;ms14,;ms17,add(19)(q13),der(19) t(?;?11;19)(?;q13;q134);ob27;cb.nuc ish 9q34(ABLx2-5) |
| 5=D | M 69 | Larynx | AML M4 | 5-FU | 2.5 | 2 | 13.2 | 219 | 0 | 210 | 5 | 4 | 0 | 0 | 0 | 4.1 | ;ms | 47,XY,;ms15,add(18)(q23),del(20) (q11q12),;plmar1,;plmar2;ob21;cb.nuc ish 9q34(ABLx2) |
| 6 | M 91 | Skin | ||||||||||||||||
| (melanoma) | ALL L1 | ;gg-Rays | 12.3 | 3 | 30.3 | 205 | 0 | 189 | 5 | 8 | 3 | 0 | 0 | 7.7 | ;ms | 45,XY,;ms7;ob12;cb/46,XY;ob9;cb.nuc ish 9q34(ABLx2) | ||
| 45,30Gy | ||||||||||||||||||
| ACNU | ||||||||||||||||||
| DTIC | ||||||||||||||||||
| 7 | M 57 | Lymphoma | AML M4 | VP-16 | 3.6 | 7 | 81.3 | 122 | 0 | 14 | 65 | 42 | 1 | 0 | 0 | 88.5 | ;ms | 45,XY,del(1)(p22),;pldel(1)(q12)x2,del(3)(p11),;ms5,;ms7, del(12)(p11),;ms16,;ms17,;ms17,;ms20,;plmar1,;plmar2, ;plmar3;ob16;cb.nuc ish 9q34(ABLx3-4) |
| Patient No. . | Sex/Age . | Primary Tumor . | Secondary Leukemia . | Therapy* . | Duration (yrs)=d . | Survival Time (mo) . | Monosomy 7 (%) . | Total Cells . | No. of ABL Gene . | ABL Gene Amplification (%) . | CD3 Amplification . | Karyotype . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | . | . | . |
| 1 | F 71 | Uterus | AML M2 | ;gg-Rays | 6.3 | 5 | 89.3 | 183 | 0 | 60 | 87 | 29 | 6 | 1 | 0 | 67.2 | ;ms | 41,XX,;ms5,;ms7,;ms15,;ms20,;ms21,;ms22,;ms22,;pl2r;ob10;cb/41,XX,;ms5.;ms7,;ms11,;ms16,;ms19,;ms20,;ms21,;plmar1, ;plmar2;ob9;cb.nuc ish 9q34(ABLx2-4) |
| 40Gy | ||||||||||||||||||
| CP | ||||||||||||||||||
| 2 | F 67 | Uterus | AML M4 | ;gg-Rays | 5.5 | 8 | 27.4 | 219 | 0 | 210 | 2 | 4 | 3 | 0 | 0 | 4.1 | ;ms | 46,XX;ob20;cb.nuc ish 9q34(ABLx2) |
| 3 | F 70 | Uterus | AML M2 | ;gg-Rays | 9.7 | 17 | 4.6 | 221 | 0 | 213 | 0 | 8 | 0 | 0 | 0 | 3.6 | ;ms | 46,XX;ob22;cb.nuc ish 9q34(ABLx2) |
| 4=D | F 63 | Breast | AML M1 | 5-FU | 7.3 | 2.3 | 5.0 | 272 | 0 | 66 | 83 | 27 | 80 | 14 | 2 | 75.7 | ;pl | 44,XX,dup(1)(q25q44),del(4)(q25q28),del(5) (q13q31),i(11)(q10),der(12)t(11;12)(q23;p12), del(13)(q12q14),;ms14,;ms17,add(19)(q13),der(19) t(?;?11;19)(?;q13;q134);ob27;cb.nuc ish 9q34(ABLx2-5) |
| 5=D | M 69 | Larynx | AML M4 | 5-FU | 2.5 | 2 | 13.2 | 219 | 0 | 210 | 5 | 4 | 0 | 0 | 0 | 4.1 | ;ms | 47,XY,;ms15,add(18)(q23),del(20) (q11q12),;plmar1,;plmar2;ob21;cb.nuc ish 9q34(ABLx2) |
| 6 | M 91 | Skin | ||||||||||||||||
| (melanoma) | ALL L1 | ;gg-Rays | 12.3 | 3 | 30.3 | 205 | 0 | 189 | 5 | 8 | 3 | 0 | 0 | 7.7 | ;ms | 45,XY,;ms7;ob12;cb/46,XY;ob9;cb.nuc ish 9q34(ABLx2) | ||
| 45,30Gy | ||||||||||||||||||
| ACNU | ||||||||||||||||||
| DTIC | ||||||||||||||||||
| 7 | M 57 | Lymphoma | AML M4 | VP-16 | 3.6 | 7 | 81.3 | 122 | 0 | 14 | 65 | 42 | 1 | 0 | 0 | 88.5 | ;ms | 45,XY,del(1)(p22),;pldel(1)(q12)x2,del(3)(p11),;ms5,;ms7, del(12)(p11),;ms16,;ms17,;ms17,;ms20,;plmar1,;plmar2, ;plmar3;ob16;cb.nuc ish 9q34(ABLx3-4) |
Therapy for the primary tumors: CP, cyclophosphamide; 5-FU, 5-fluorouracil; ACNU, nimustine; DTIC, dacarbazine; VP-16, etoposide.
=d Interval from last therapy till diagnosis of secondary leukemia.
=D Hiroshima A-bomb survivors.
RESULTS
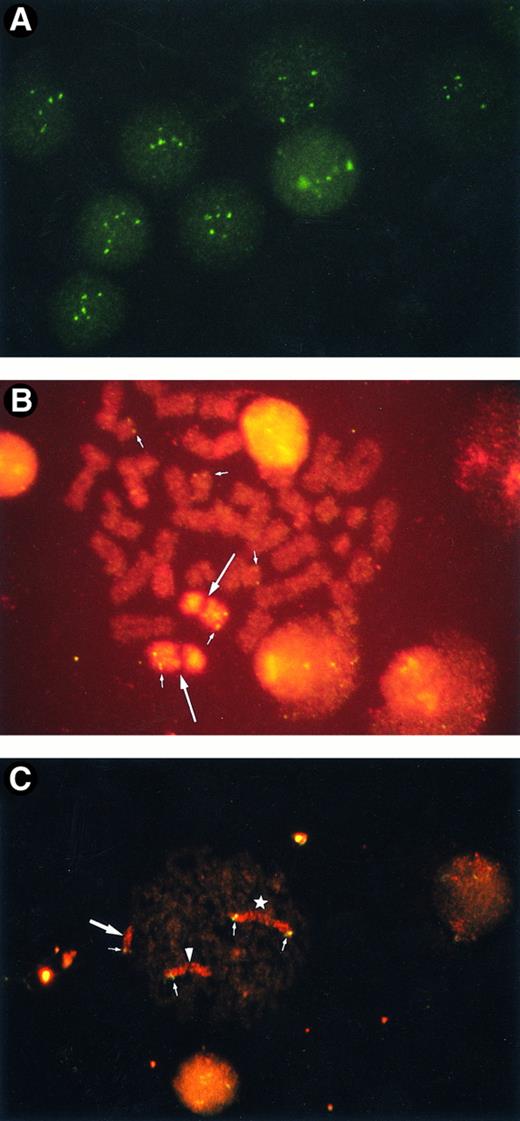
Amplification of ABL oncogene.FISH analysis using the ABL oncogene probe revealed that 3 of the 7 patients with secondary leukemia (42.9%) had an amplified ABL oncogene. Patients no. 1, 4, and 7 had 67.2%, 75.7%, and 88.5% of the interphase nuclei containing more than three signals, respectively (Table 1). Representative interphase nuclei with four to six ABL signals are shown in Fig 1A.
(A) Amplified ABL oncogenes in patient no. 4, showing four to six signals in interphase cells. Some signals are slightly out of focus. (B) Amplified ABL oncogenes in patient no. 1, representing five copies in one metaphase (small arrows). Two homologues of chromosome 9 are stained with a spectrum orange WCP9 probe (large arrows). (C) Amplified chromosomal segment containing CD3-MLL genes from patient no. 4. Four chromosomal segments of 11q23 were found in one metaphase (small arrows): one was on the normal chromosome 11 (arrowhead), one on a derivative 12 (large arrow), and two on the isochromosome 11q10 (star). Chromosome 11 is painted in red with rhodamine and the CD3 gene in green with fluorescein.
(A) Amplified ABL oncogenes in patient no. 4, showing four to six signals in interphase cells. Some signals are slightly out of focus. (B) Amplified ABL oncogenes in patient no. 1, representing five copies in one metaphase (small arrows). Two homologues of chromosome 9 are stained with a spectrum orange WCP9 probe (large arrows). (C) Amplified chromosomal segment containing CD3-MLL genes from patient no. 4. Four chromosomal segments of 11q23 were found in one metaphase (small arrows): one was on the normal chromosome 11 (arrowhead), one on a derivative 12 (large arrow), and two on the isochromosome 11q10 (star). Chromosome 11 is painted in red with rhodamine and the CD3 gene in green with fluorescein.
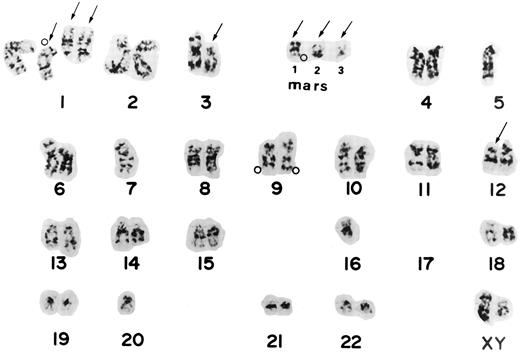
The ABL oncogene was also examined on chromosomes from the 3 patients. In patient no. 1, two ABL signals were located on homologous chromosome 9 and three on marker chromosomes (Fig 1B). Patient no. 4 had one to three extra ABL oncogene signals on the terminal portions of derivative chromosomes. Patient no. 7 had three to four ABL signals. The ABL signals were observed in a derivative chromosome 1 and in one marker chromosome besides both normal chromosomes 9. A representative karyotype with four ABL oncogene signals from patient no. 7 is shown in Fig 2. Thus, the region involving the ABL oncogene moved to several other abnormal chromosomes, resulting in partial tetra-, penta-somy of the 9q34 region. A minisatellite cosmid probe (D9S7), located on the 9q34 region near the ABL oncogene, also translocated to other chromosomes, to which the ABL oncogene moved. DNAs from these 3 patients were Southern blotted using a cDNA ABL probe (Oncor Science). They showed a single amplified germ line band, indicating that there was no gross alteration of the ABL oncogene. These results suggested that the 9q34 region translocated to other chromosomes as “a segment.”
Karyotype from patient 7 shows the complex karyotype, -4 5 , X Y , d e l ( 1 ) ( p 2 2 ) , ;k1 d e l ( 1 ) ( q 1 2 ) ;k4 2 , d e l ( 3 ) ( p 1 1 ) , ;k2 5 , ;k2 7 , d e l ( 1 2 ) ( p 1 1 ) , ;k2 1 6 , ;k2 1 7 , ;k2 1 7 , ;k2 2 0 , ;k1 m a r 1 , ;k1mar2,;k1mar3. Arrows show chromosomes with structural aberrations. Open circles (;pe) indicate the locations of the ABL oncogene detected by FISH from the same Carnoy's solution. Two signals were located on two homologues of chromosome 9, one on derivative chromosome 1, and one on a marker chromosome. Mars corresponds to marker chromosomes.
Karyotype from patient 7 shows the complex karyotype, -4 5 , X Y , d e l ( 1 ) ( p 2 2 ) , ;k1 d e l ( 1 ) ( q 1 2 ) ;k4 2 , d e l ( 3 ) ( p 1 1 ) , ;k2 5 , ;k2 7 , d e l ( 1 2 ) ( p 1 1 ) , ;k2 1 6 , ;k2 1 7 , ;k2 1 7 , ;k2 2 0 , ;k1 m a r 1 , ;k1mar2,;k1mar3. Arrows show chromosomes with structural aberrations. Open circles (;pe) indicate the locations of the ABL oncogene detected by FISH from the same Carnoy's solution. Two signals were located on two homologues of chromosome 9, one on derivative chromosome 1, and one on a marker chromosome. Mars corresponds to marker chromosomes.
Segmental jumping translocations (SJT) of the ABL oncogene also were examined in 15 de novo AML patients, including 5 with complex chromosome aberrations, 5 with specific chromosome abnormalities of t(9; 22), t(8; 21), t(15; 17), 11q23 translocations or inv(16), and 5 with a normal karyotype (Table 2). None of them had an amplified ABL oncogene with two ABL signals on homologous chromosome 9. Twenty patients with chronic myelocytic leukemia (CML) in the chronic phase, 12 with MDS, and 7 individuals without hematologic diseases, did not have an amplified ABL oncogene. The SJT of the ABL oncogene was essentially restricted to secondary leukemias.
SJT of ABL Oncogene Found in Leukemia Patients and Normal Individuals
| Diseases . | No. of Patients Observed . | No. of Positive Patients . |
|---|---|---|
| Secondary leukemia | 7 | 3 |
| De novo AML | 15 | 0 |
| CML (chronic phase) | 20 | 0 |
| MDS | 12 | 0 |
| Individuals without hematologic disease | 7 | 0 |
| Diseases . | No. of Patients Observed . | No. of Positive Patients . |
|---|---|---|
| Secondary leukemia | 7 | 3 |
| De novo AML | 15 | 0 |
| CML (chronic phase) | 20 | 0 |
| MDS | 12 | 0 |
| Individuals without hematologic disease | 7 | 0 |
Analysis of the 11q23 region on chromosome 11.The 11q23 region was also analyzed in the same samples from these seven secondary leukemia patients by means of FISH on metaphases and on interphase nuclei, using mixtures of a whole chromosome painting probe for chromosome 11 and a CD3 YAC probe that contains the CD3 and MLL genes. Of the 7 patients, 1 (patient no. 4) had an amplified 11q23 region. This patient had an isochromosome of the long arm of chromosome 11, i(11)(q10) and a derivative chromosome 12. FISH revealed that the patient had a total of four CD3-MLL signals on both metaphases and on interphase nuclei. Iso chromosome 11 had two signals. The terminal portion of the derivative chromosome 12 was stained by whole chromosome painting probe for chromosome 11, on which CD3 YAC was located. A representative metaphase analyzed by FISH is shown in Fig 1C.
As shown in Table 1, 4 of the 7 secondary leukemia patients (patients no. 1, 4, 5, and 7) had complex karyotypes, involving monosomy or a deletion of chromosomes 5 or 7. Of the 4 patients, 3 had an SJT of ABL alone or in combination with CD3 oncogenes.
Analysis of monosomy 7.The incidence of monosomy 7-positive cells in the 7 secondary leukemia patients was investigated by interphase FISH. Monosomy 7-positive cells were found in 5 (patients no. 1, 2, 5, 6, and 7) of the 7 patients (71.4%). The frequency of monosomy 7 cells in these patients was 89.3%, 27.4%, 13.2%, 30.3%, and 81.3%, respectively, in 500 nuclei scored (Table 1). Of the 5 patients with monosomy 7, 2 also had ABL oncogene amplification. Patient no. 1 was followed up for 2 years. During that period, the frequency of the monosomy 7 increased from 14.1% to 89.3%.
DISCUSSION
The present study showed that the ABL oncogene alone or in combination with CD3-MLL regions was frequently amplified in secondary leukemia. The amplified genes were located on structurally abnormal chromosomes, especially marker chromosomes. Monosomy 7 was also found in patients with the gene amplification. Our investigation showed that a specific chromosomal segment moves onto several other chromosomes, resulting in partial tri-, tetra-, or penta-somy. Southern blotting revealed no gene rearrangement of the ABL oncogene itself. Therefore, a chromosomal segment of 9q34 involving ABL gene, as a unit, translocated onto other chromosomes. A similar SJT was found in the 11q23 region of chromosome 11, where the CD3 and MLL genes are located.
In conventional chromosome analysis, “jumping translocation” has been identified. However, only ten instances have been reported.13 14 Part of the long arm of chromosome 1 was found on several other chromosomes as a result of a jumping translocation. Because the translocated chromosome has typical band patterns, it was easily detected. However, smaller parts of translocated regions were virtually undetectable by conventional chromosome analysis. FISH using whole chromosome painting and specific oncogene probes will be very helpful in identifying these segmentally translocated regions.
A FISH mapping study of micro dissected chromosome regions obtained from HSRs in nine breast cancer cell lines showed that the HSRs are composed of integrated amplified chromosome regions derived from two or more chromosome regions.15
In our study, 4 patients were treated with a combination of extensive chemo- and radiation-therapy; 1 patient was treated with etoposide. Therapy for malignant lymphomas and solid tumors has become more extensive, combining drugs such as etoposide, daunorubicin, and 6-mercaptopurine, with irradiation. The mechanism for the malignant transformation by etoposide may differ from that of alkylating agents due to the different molecular pharmacology of these drugs. Etoposide interacts with DNA by blocking the catenative activity of topoisomerase II inhibitors, leading to DNA strand breakage, which results in a high frequency of sister chromatid exchanges (SCE) and other chromosome aberrations.16 17
The precise mechanisms for the SJTs remain unknown. Several mechanisms can be proposed. The first is that these chromosomal regions cannot control the replication of amplifying DNA sequences, which leads to recombinations. The second is that the chromosomal segments may be deleted from the original site and form an episome, resulting in gene amplification. The HL-60 cell line at passages 46 to 62 contained amplified c-MYC sequences on submicroscopic circular extrachromosomal DNAs called episomes.18,19 With increased passages, the HL-60 cells lost the episome c-MYC sequences and shifted to dmin. Over time, the c-MYC sequences moved from their extra chromosomal sites onto the short arm of the derivative chromosome 5. The episome is usually only 500 to 900 kb long in the HL-60 cell line.19 An SJT with ABL oncogene contained approximately half of the chromosomal band consisting of 10 to 100 Mb. Therefore, the mechanism of the segmental jumping translocation seems to differ from that of the episome. Other mechanisms that explain how large chromosomal regions move to other chromosomal regions remain to be resolved. The third possibility is that unequal mitotic nonhomologous chromosomal recombination might occur among highly unstable chromosomal and hyper replication sites. There is some evidence that HSRs are generated by unequal crossing-over between chromatids with similar DNA structures.20
The present study indicates that the SJT of ABL and/or CD3 (MLL) genes is associated with the leukemogenesis of secondary leukemia, and that the SJT is one mechanism of gene amplification.
ACKNOWLEDGMENT
We wish to thank Dr M. Seto (Aichi Cancer Center, Japan) for providing the CD3 YAC probe.
Supported in part by Grants-in-Aid for General Scientific Research (07671208), for Scientific Research on Priority Areas (06282114) from the Ministry of Education, Science and Culture, the Research Fund for Comprehensive Cancer Treatment (1995), and the Research Fund of the Sagawa Foundation (1995).
Address reprint requests to Nanao Kamada, MD, Department of Cancer Cytogenetics, Research Institute for Radiation Biology and Medicine, Hiroshima University, 1-2-3, Kasumi, Minami-ku, Hiroshima 734, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal