To the Editor:
Recent studies have shown that structural abnormalities (both deletions and translocations) involving chromosome 13 are frequent in chronic lymphocytic leukemia (CLL), occurring in about 17% to 29% of patients.1-3 Deletions of varying length almost always involve the q14 band (75% of samples), of which the most common are 13q12-14 and 13q14-22. The search for a candidate tumor-suppressor gene on 13q14 has been recently restricted to a region containing the band 13q14.3.4-9 Using fluorescence in situ hybridization (FISH) and/or Southern blotting analysis,4-9 the minimum area of deletion has been shown to be between the RB1 and D13S3194 5 centromeric to the D13S25 marker.
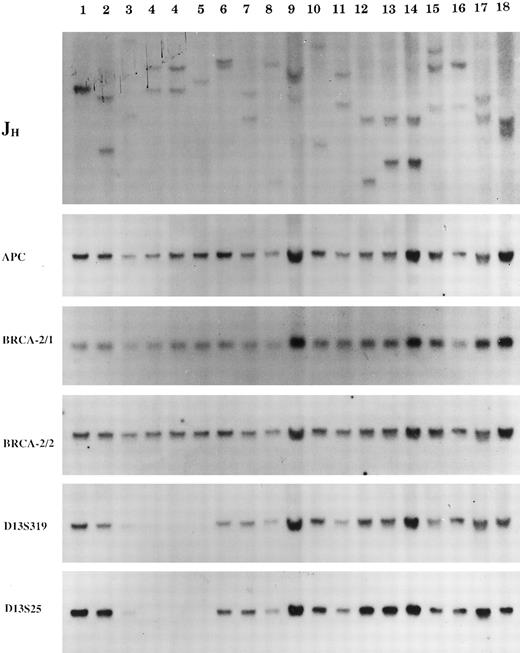
Representative examples of Southern blot analysis of CLL patients. A panel of probes were used for the investigation of clonality (IgH joining region probe), control or loading of total DNA (APC probe), deletion of 13q14.3 region (D13S25 and D13S319 marker probes), and deletion of 13q12 (BRCA2 probe for exon 11, BRCA2-1 and exon 26-27, BRCA2-2, respectively). Except for the patients in lanes 9, 15, and 16 (with <90% clonal B cells population as determined by the JH pattern), all of the other patients are representative examples of patterns included in this study.
Representative examples of Southern blot analysis of CLL patients. A panel of probes were used for the investigation of clonality (IgH joining region probe), control or loading of total DNA (APC probe), deletion of 13q14.3 region (D13S25 and D13S319 marker probes), and deletion of 13q12 (BRCA2 probe for exon 11, BRCA2-1 and exon 26-27, BRCA2-2, respectively). Except for the patients in lanes 9, 15, and 16 (with <90% clonal B cells population as determined by the JH pattern), all of the other patients are representative examples of patterns included in this study.
More recently, Garcia Marco et al10 have proposed that a more centromeric region on chromosome 13 at band 13q12, the site of the breast cancer susceptibility gene 2 (BRCA2) at 13q12.2,11 is the site of the major deletion occurring in CLL cells. Using FISH analysis, they found a varying percentage of cells (from 13% to 95%) to carry such a deletion in 80% of CLL patients, which implies that this is the most common chromosomal abnormality in CLL. Both homozygous and heterozygous cases have been reported in their cohort of 35 patients studied.10
To investigate the involvement of the 13q12 and 13q14 region, we have analyzed 44 B-CLL patients and tested by Southern blotting for deletion of the D13S25 and D13S319 markers (from 13q14.3) and for deletion of the BRCA2 gene (probes for exon 11 and 26-27). Signals obtained by hybridization were measured by densitometry and intensity was compared with the intensity of a control sequence for the adenomatous polyposis coli gene (APC) from chromosome 5q31 or the T-cell receptor δ (TCRD) gene on chromosome 14q11.2, which have not been previously found to be affected by chromosomal abnormalities in CLL.2 Patients were tested for deletion of the D13S25 (44 patients), D13S319 (27 patients), and BRCA2 (24 patients). Only patients with ≥90% clonal B cells (as measured using hybridization to the JH probe) were assessed for 13q14 deletion (see Fig 1, top panel and legend). Homozygous and heterozygous deletion of the D13S25 was detected in 5 and 17 patients, respectively (average ratio of D13S25/APC, 0.03 and 0.533), whereas 22 patients showed no deletion (average ratio, 1.003). Representative examples of the patterns obtained are shown in Fig 1. Twenty-seven of these patients were tested with the D13S319 marker and showed a pattern identical to that detected by the D13S25. In contrast to the data obtained using the 13q14.3 probes, the use of two separate probes from the BRCA2 gene failed to detect clonal homozygous or heterozygous deletion in any of 24 CLL patients tested (average ratio of BRCA2/APC or BRCA2/TCRD, 1.04), including 4 patients with homozygous deletion of D13S25 and D13S319 (Fig 1).
In conclusion, our study indicates that deletion involving the D13S25 and D13S319 occurs in 48% of CLL patients studied. Because the DNA used was derived from a clonal population, we conclude that the 13q14.3 deletion is a clonal event in the CLL patients analyzed. By contrast, no homozygous or heterozygous clonal deletion was detected by either probes for the BRCA2 gene in any of the patients studied.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal