Abstract
To clarify whether the expression of the WT1 gene in leukemic cells is aberrant or merely reflects that in normal counterparts, the expression levels of the WT1 gene were quantitated for normal hematopoietic progenitor cells. Bone marrow (BM) and umbilical cord blood (CB) cells were fluorescence-activated cell sorting (FACS)-sorted into CD34+ and CD34− cell populations, and the CD34+ cells into nine subsets (CD34+CD33−, CD34+CD33+, CD34+CD38−, CD34+CD38+, CD34+HLA-DR−, CD34+HLA-DR+, CD34+c-kithigh, CD34+c-kitlow, and CD34+c-kit−) according to the expression levels of CD34, CD33, CD38, HLA-DR, and c-kit. Moreover, acute myeloid leukemic cells were also FACS-sorted into four populations (CD34+CD33−, CD34+CD33+, CD34− CD33+, and CD34− CD33−). FACS-sorted normal hematopoietic progenitor and leukemic cells and FACS-unsorted leukemic cells were examined for the WT1 expression by quantitative reverse transcriptase-polymerase chain reaction. The WT1 expression in the CD34+ and CD34− cell populations and in the nine CD34+ subsets of BM and CB was at either very low (1.0 to 2.4 × 10−2) or undetectable (<10−2) levels (the WT1 expression level of K562 cells was defined as 1.0), whereas the average levels of WT1 expression in FACS-sorted and -unsorted leukemic cells were 2.4 to 9.3 × 10−1. Thus, the WT1 expression levels in normal hematopoietic progenitor cells were at least 10 times less than those in leukemic cells. Therefore, we could not find any normal counterparts of BM or CB that expressed the WT1 at levels comparable with those in leukemic cells. These results indicate an aberrant overexpression of the WT1 gene in leukemic cells and imply the involvement of this gene in human leukemogenesis.
UNLIKE TUMOR SUPPRESSOR genes Rb and p53, which are expressed ubiquitously, the expression of the WT1 gene is restricted to a limited set of tissues such as fetal kidney, ovary, testis, spleen, and the mesothelial cell lining of visceral organs.1,2 Analysis of mice carrying disrupted WT1 gene showed a crucial role of the WT1 gene in early urogenital development.3 In the hematopoietic system, WT1 mRNA was detected at high levels in spleen and at low levels (10−4; the WT1 expression level in K562 cells was defined as 1.0) in bone marrow (BM). WT1 expression in spleen was found to be restricted to the splenic capsule and stroma, but was not detected in splenocytes.2 However, little is known about WT1 expression in normal hematopoietic progenitor cells.
Concerning the involvement of the WT1 gene in human malignancies other than Wilms tumor, it has been reported that the WT1 gene was mutated in a human mesothelioma2 and that some human leukemia samples expressed the WT1 gene.4 We recently found that all 96 human leukemia samples examined by us expressed the WT1 gene, regardless of the disease subtype (acute myelogenous leukemia [AML], acute lymphocytic leukemia [ALL], or chronic myelogenous leukemia [CML]) as a result of raising the detection sensitivity of WT1 mRNA by means of reverse transcriptase-polymerase chain reaction (RT-PCR) assay.5 We showed in the same study that the WT1 expression levels increased in parallel with the progression of clinical stage in CML and correlated inversely with the prognosis for acute leukemia.5 Thus, WT1 was identified as a new prognostic factor and a new marker for the diagnosis of minimal residual disease in human leukemia.5,6 Subsequently, we found that WT1 antisense oligomers inhibited the growth of fresh human leukemic cells and leukemic cell lines, but not that of normal colony-forming unit granulocyte-macrophage (CFU-GM), suggesting an important role of WT1 in leukemic cell growth.7 However, it is unknown whether the WT1 expression in leukemic cells is aberrant or merely reflects that in normal counterparts.
In the present study, we examined the expression levels of the WT1 gene in fluorescence-activated cell sorting (FACS)-sorted hematopoietic progenitor cells of BM and cord blood (CB) to determine whether or not the WT1 overexpression in leukemic cells is aberrant. We describe here that the WT1 expression in normal hematopoietic progenitor cells of BM and CB is either very low or undetectable and at least 10 times less than that in leukemic cells. The role of the WT1 gene in human leukemogenesis is also discussed.
MATERIALS AND METHODS
Cell lines.SAM-1 and BV173 (derived from CML, blastic crisis) were kindly provided by Dr E. Tatsumi (Kobe University, Hyogo, Japan) and Dr J. Minowada (Hayashibara Biochemical Labs, Inc, Okayama, Japan), respectively. Kasumi-1 (derived from t[8; 21] AML-M2), was kindly provided by Dr K. Kita (Mie University, Mie, Japan), TF-1 (derived from AML-M6) by Dr T. Kitamura (DNAX Research Institute, Palo Alto, CA), DL-40 (derived from non-Hodgkin's lymphoma, diffuse large-cell type) by Dr I. Kubanishi (Kochi Medical School, Kochi, Japan), and KT-3 (interleukin-6 [IL-6]–dependent T-cell lymphoma cell line) by Dr S. Shimizu (Kanazawa Medical University, Ishikawa, Japan). The remaining cell lines were kindly provided by the Japanese Cancer Research Resources Bank (Tokyo, Japan). The cell lines were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (FBS).
Cell preparation.After informed consent was obtained, BM cells were collected by aspiration from the posterior iliac crest of healthy adult volunteers. Umbilical CB was obtained after normal vaginal deliveries at full term by aspiration from pregnant women from whom informed consent was also obtained. BM aspirate was placed in a 16-mL Falcon tissue culture tube and CB in a 50-mL one containing 400 U of preservative-free heparin (Shimizu Pharmaceutical Co, Osaka, Japan). Mononuclear cells were separated by using Ficoll-Paque (Pharmacia, Piscataway, NJ) density gradient centrifugation, and nonadherent cells were recovered by overnight adherence to plastic dishes. The mononuclear nonadherent cell fractions were further enriched by negative selection using soybean agglutinin (SBA) Micro-CELLector Flasks (Applied Immune Science Inc, Menlo Park, CA) according to the manufacturer's instructions. Leukemic cells were isolated by aspiration from BM or peripheral blood (PB) of patients with leukemia, and their mononuclear cell fractions were obtained as described above. For some of the experiments, previously frozen mononuclear cells were used after rapid thawing.
Purification of hematopoietic progenitor and leukemic cells by cell sorting.Highly purified progenitors were separated from BM- or CB- derived SBA− cells by using a FACStar Plus (Becton Dickinson [BD] Immunocytometry Systems, San Jose, CA) equipped with an argon laser tuned at 488 nm, as reported elsewhere.8,9 BM- or CB-derived SBA− cells were washed twice with phosphate-buffered saline (PBS)(-) containing 2% FBS (staining medium) and were passed through a stainless-steel mesh. The cells were then pelleted before staining with monoclonal antibodies (MoAbs). The following MoAbs were used: fluorescein isothiocyanate (FITC)-conjugated HPCA-2 (mouse IgG1 CD34 MoAb; BD); phycoerythrin (PE)-conjugated HLA-DR (mouse IgG2a; BD); PE-conjugated CD33 (MY9; mouse IgG2b; Coulter Immunology, Hialeah, FL); PE-conjugated CD38 (mouse IgG1; BD); purified antihuman c-kit MoAb (mouse IgM; Immunotech S.A., Marseille Cedex, France); and goat antimouse IgM with PE (Immunotech S.A.). For double staining, cells were incubated with 20 μL of HLA-DR, CD33, or CD38/106 cells for 30 minutes on ice and then washed twice with staining medium. Cells were also stained with 20 μL of purified antihuman c-kit MoAb/5 × 105 cells for 30 minutes at room temperature. After washing with staining medium, cells were incubated with 20 μL of goat antimouse IgM with PE/5 × 105 cells for 30 minutes at room temperature. These cells were then washed twice with staining medium and stained with HPCA-2 (FITC) as described elsewhere.8 9 All the stained cells were kept on ice until cell sorting. For negative controls, unstained cells and cells stained only with second antibody or with isotype control IgG with either FITC or PE were included.
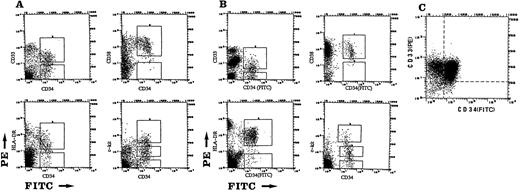
Stained cells were sorted with a FACStar Plus single-laser flow cytometry system. Sorting gates were established for both forward scattering and side scattering, and dual-parameter dotograms showing FITC (CD34) and PE (CD33, CD38, HLA-DR, or c-kit) fluorescence were generated from the gated events. These gated dotograms were then used to sort the cells into CD34+CD33+/−, CD34+CD38+/−, or CD34+HLA-DR+/− cells. The sorting windows for CD34+ cells expressing different levels of c-kit protein were also established as previously reported.9 CD34+c-kithigh, low or − cells were then separately sorted. Typical scattergrams of BM-, CB-, or leukemia-derived CD34+ cells expressing different levels of CD33, CD38, HLA-DR, or c-kit are presented in Figs 1, 2, and 3. Data were aquired with FACStar Plus Research Software, and a minimum of 20,000 events was analyzed for each sample. After sorting, the recovered cells were washed twice with α-medium, then washed with ice-cold PBS(-) and stored in D solution at −80°C for the detection of WT1 mRNA by RT-PCR. The purity of FACS-sorted cells of BM and CB was greater than 90%.
Expression of CD33, CD38, HLA-DR, or c-kit on BM- derived CD34+ cells (A) or CB-derived CD34+ cells (B). Sorting gates were first established for forward scattering and side scattering. Four dual-parameter dotograms displaying FITC (CD34) and PE (CD33, CD38, HLA-DR, or c-kit) fluorescence were then generated from the gated events. (C) Leukemic cells obtained from a patient with AML (M4, patient no. 294) were sorted as described. A dual-parameter dotogram displaying CD34 and CD33 is shown.
Expression of CD33, CD38, HLA-DR, or c-kit on BM- derived CD34+ cells (A) or CB-derived CD34+ cells (B). Sorting gates were first established for forward scattering and side scattering. Four dual-parameter dotograms displaying FITC (CD34) and PE (CD33, CD38, HLA-DR, or c-kit) fluorescence were then generated from the gated events. (C) Leukemic cells obtained from a patient with AML (M4, patient no. 294) were sorted as described. A dual-parameter dotogram displaying CD34 and CD33 is shown.
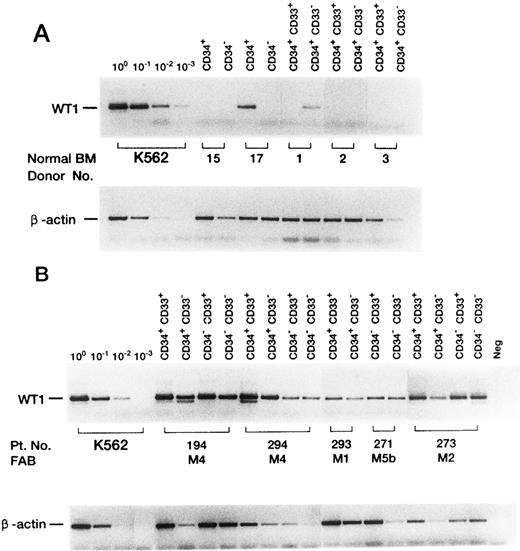
Expression of the WT1 gene in FACS-sorted normal BM (A) or leukemic (B) cells. Representative cases are shown. To detect WT1 mRNA, PCR was performed for 45 cycles (35 and 10 cycles for first and second round PCR, respectively). To quantitate β-actin mRNA, PCR was performed for 31 cycles.
Expression of the WT1 gene in FACS-sorted normal BM (A) or leukemic (B) cells. Representative cases are shown. To detect WT1 mRNA, PCR was performed for 45 cycles (35 and 10 cycles for first and second round PCR, respectively). To quantitate β-actin mRNA, PCR was performed for 31 cycles.
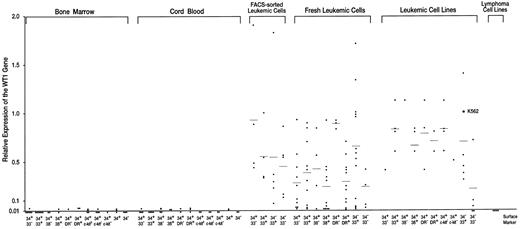
Expression levels of the WT1 gene in normal hematopoietic progenitor and leukemic cells. Expression levels of the WT1 gene in K562 cells were defined as 1.0.
Expression levels of the WT1 gene in normal hematopoietic progenitor and leukemic cells. Expression levels of the WT1 gene in K562 cells were defined as 1.0.
RT-PCR.Quantitative RT-PCR of FACS-sorted cells was performed as described previously5,10 with some modifications. Briefly, total RNA was extracted from 2,000 to 4,000 cells with the acid guanidine-phenol chloroform method and then converted into cDNA on a scale of 20 μL. The expression level of the WT1 gene in K562 cells was defined as 1.0 as described previously.5 PCR was performed for 35 cycles with the outer primers to quantitate 100 to 10−2 levels of WT1 expression. For the quantitation of 10−2 to 10−3 levels of the WT1 expression, a second PCR using nested internal primers was performed for 10 to 14 cycles. To quantitate β-actin levels, PCR was also performed for 30 to 32 cycles. To normalize the differences in RNA degradation for individual samples and in RNA loading for RT-PCR, the value of the WT1 gene expression divided by the β-actin gene expression was defined as the WT1 expression levels of the samples. Serial 1:10 dilutions of standard cDNAs prepared from 2,000 K562 cells were always PCR-amplified simultaneously with the samples to determine the WT1 expression levels of the samples relative to those of K562 cells. Because the amount of total RNA that could be extracted was very small because of the small number of FACS-sorted cells, the detection limit of WT1 mRNA by RT-PCR was restricted to the 10−2 level. Quantitative RT-PCR of fresh leukemic cells and leukemic cell lines was performed as described previously5 10 with some modifications. Briefly, 2.0 μg of total RNA from leukemic samples was converted into cDNA in 30 μL of reaction buffer. PCR was performed for 27 to 30 cycles for the quantitation of WT1 mRNA and for 18 cycles for the quantitation of β-actin mRNA. All experiments were performed in duplicate.
RESULTS
Very low or undetectable levels of WT1 expression in hematopoietic progenitor cells of BM and umbilical CB.BM and umbilical CB cells were FACS-sorted according to the levels of the expression of CD34, CD33, CD38, HLA-DR, and c-kit (Figs 1A and B). CD34+ and CD34− cell populations, and nine CD34+ subsets of CD34+CD33−, CD34+CD33+, CD34+CD38−, CD34+CD38+, CD34+HLA-DR−, CD34+HLA-DR+, CD34+c-kithigh, CD34+c-kitlow, and CD34+ c-kit− were isolated and examined for the expression levels of the WT1 gene (Figs 2 and 3, Table 1). The WT1 expression levels of CD34+ and CD34− cell populations and of the nine CD34+ subsets of BM and CB were all very low (1.0 to 2.4 × 10−2) or undetectable (<10−2). Of 5 CD34+ cell samples (3 and 2 samples from BM and CB, respectively), 4 expressed very low levels (1.0 to 2.4 × 10−2) of WT1, whereas the WT1 expression in all the 5 CD34− cell samples (3 and 2 samples from BM and CB, respectively) were at undetectable levels (<10−2). Thus, the WT1 expression levels were significantly (P < .01) higher in CD34+ cells than in CD34− cells. No significant difference in the WT1 expression levels was observed among nine different CD34+ subsets (28 and 27 samples from BM and CB, respectively) under the present experimental conditions, where detection limit of the WT1 expression was at 10−2 levels.
Expression of WT1 mRNA in FACS-Sorted Cells
| BM | CB | ||||
| Donor No. | CD34+CD33− | CD34+CD33+ | Donor No. | CD34+CD33− | CD34+CD33+ |
| 1 | 2.0 × 10−2 | <10−2 | 18 | <10−2 | <10−2 |
| 2 | <10−2 | <10−2 | 19 | <10−2 | <10−2 |
| 3 | <10−2 | <10−2 | 20 | <10−2 | <10−2 |
| 4 | <10−2 | <10−2 | 21 | 1.5 × 10−2 | <10−2 |
| 5 | <10−2 | <10−2 | |||
| 6 | <10−2 | <10−2 | |||
| BM | CB | ||||
| Donor No. | CD34+CD33− | CD34+CD33+ | Donor No. | CD34+CD33− | CD34+CD33+ |
| 1 | 2.0 × 10−2 | <10−2 | 18 | <10−2 | <10−2 |
| 2 | <10−2 | <10−2 | 19 | <10−2 | <10−2 |
| 3 | <10−2 | <10−2 | 20 | <10−2 | <10−2 |
| 4 | <10−2 | <10−2 | 21 | 1.5 × 10−2 | <10−2 |
| 5 | <10−2 | <10−2 | |||
| 6 | <10−2 | <10−2 | |||
| Donor No. | CD34+CD38− | CD34+CD38+ | Donor No. | CD34+CD38− | CD34+CD38+ |
| 7 | <10−2 | <10−2 | 22 | <10−2 | <10−2 |
| 8 | <10−2 | 1.3 × 10−2 | 23 | <10−2 | 1.0 × 10−2 |
| 24 | 1.3 × 10−2 | <10−2 | |||
| 25 | 1.7 × 10−2 | 1.0 × 10−2 | |||
| Donor No. | CD34+CD38− | CD34+CD38+ | Donor No. | CD34+CD38− | CD34+CD38+ |
| 7 | <10−2 | <10−2 | 22 | <10−2 | <10−2 |
| 8 | <10−2 | 1.3 × 10−2 | 23 | <10−2 | 1.0 × 10−2 |
| 24 | 1.3 × 10−2 | <10−2 | |||
| 25 | 1.7 × 10−2 | 1.0 × 10−2 | |||
| Donor No. | CD34+HLA-DR− | CD34+HLA-DR+ | Donor No. | CD34+HLA-DR− | CD34+HLA-DR+ |
| 9 | 1.3 × 10−2 | 2.2 × 10−2 | 26 | NE | <10−2 |
| 10 | NE | 2.4 × 10−2 | 27 | <10−2 | 1.0 × 10−2 |
| 28 | 1.4 × 10−2 | 2.4 × 10−2 | |||
| Donor No. | CD34+HLA-DR− | CD34+HLA-DR+ | Donor No. | CD34+HLA-DR− | CD34+HLA-DR+ |
| 9 | 1.3 × 10−2 | 2.2 × 10−2 | 26 | NE | <10−2 |
| 10 | NE | 2.4 × 10−2 | 27 | <10−2 | 1.0 × 10−2 |
| 28 | 1.4 × 10−2 | 2.4 × 10−2 | |||
| Donor No. | CD34+c-kithigh | CD34+c-kitlow | CD34+c-kit− | Donor No. | CD34+c-kithigh | CD34+c-kitlow | CD34+c-kit− |
| 11 | <10−2 | <10−2 | <10−2 | 29 | <10−2 | <10−2 | <10−2 |
| 12 | <10−2 | <10−2 | NE | 30 | <10−2 | <10−2 | 1.0 × 10−2 |
| 13 | 1.3 × 10−2 | <10−2 | NE | ||||
| 14 | 1.0 × 10−2 | 1.5 × 10−2 | NE | ||||
| Donor No. | CD34+c-kithigh | CD34+c-kitlow | CD34+c-kit− | Donor No. | CD34+c-kithigh | CD34+c-kitlow | CD34+c-kit− |
| 11 | <10−2 | <10−2 | <10−2 | 29 | <10−2 | <10−2 | <10−2 |
| 12 | <10−2 | <10−2 | NE | 30 | <10−2 | <10−2 | 1.0 × 10−2 |
| 13 | 1.3 × 10−2 | <10−2 | NE | ||||
| 14 | 1.0 × 10−2 | 1.5 × 10−2 | NE | ||||
| Donor No. | CD34+ | CD34− | Donor No. | CD34+ | CD34− |
| 15 | <10−2 | <10−2 | 31 | 1.5 × 10−2 | <10−2 |
| 16 | 1.0 × 10−2 | <10−2 | 32 | 2.4 × 10−2 | <10−2 |
| 17 | 2.2 × 10−2 | <10−2 | |||
| Donor No. | CD34+ | CD34− | Donor No. | CD34+ | CD34− |
| 15 | <10−2 | <10−2 | 31 | 1.5 × 10−2 | <10−2 |
| 16 | 1.0 × 10−2 | <10−2 | 32 | 2.4 × 10−2 | <10−2 |
| 17 | 2.2 × 10−2 | <10−2 | |||
| FACS-Sorted Leukemic Cells . | |||||||
|---|---|---|---|---|---|---|---|
| Patient No. FAB . | CD34 (%) . | CD33 (%) . | Unsorted . | CD34+CD33+ . | CD34+CD33− . | CD34−CD33+ . | CD34−CD33− . |
| 293 | 68 | 58 | 1.5 × 10−1 | 5.4 × 10−1 | 4.8 × 10−1 | 2.9 × 10−1 | 1.6 × 10−1 |
| M1 | |||||||
| 273 | 79 | 92 | 8.9 × 10−1 | 9.9 × 10−1 | 1.9 × 100 | 1.8 × 100 | 5.6 × 10−1 |
| M2 | |||||||
| 194 | 10 | 72 | 6.4 × 10−1 | 3.4 × 10−1 | 8.9 × 10−1 | 3.6 × 10−1 | 8.6 × 10−1 |
| M4 | |||||||
| 294 | 78 | 96 | 4.1 × 10−1 | 3.5 × 10−1 | 4.4 × 10−1 | 2.3 × 10−1 | 5.4 × 10−1 |
| M4 | |||||||
| 271 | 9 | 91 | 1.0 × 10−2 | ND | ND | 7.0 × 10−2 | 1.3 × 10−1 |
| M5b | |||||||
| FACS-Sorted Leukemic Cells . | |||||||
|---|---|---|---|---|---|---|---|
| Patient No. FAB . | CD34 (%) . | CD33 (%) . | Unsorted . | CD34+CD33+ . | CD34+CD33− . | CD34−CD33+ . | CD34−CD33− . |
| 293 | 68 | 58 | 1.5 × 10−1 | 5.4 × 10−1 | 4.8 × 10−1 | 2.9 × 10−1 | 1.6 × 10−1 |
| M1 | |||||||
| 273 | 79 | 92 | 8.9 × 10−1 | 9.9 × 10−1 | 1.9 × 100 | 1.8 × 100 | 5.6 × 10−1 |
| M2 | |||||||
| 194 | 10 | 72 | 6.4 × 10−1 | 3.4 × 10−1 | 8.9 × 10−1 | 3.6 × 10−1 | 8.6 × 10−1 |
| M4 | |||||||
| 294 | 78 | 96 | 4.1 × 10−1 | 3.5 × 10−1 | 4.4 × 10−1 | 2.3 × 10−1 | 5.4 × 10−1 |
| M4 | |||||||
| 271 | 9 | 91 | 1.0 × 10−2 | ND | ND | 7.0 × 10−2 | 1.3 × 10−1 |
| M5b | |||||||
Abbreviations: NE, not able to evaluate because of low levels of β-actin; ND, not determined; FAB, French-American-British criteria.
High levels of WT1 expression in leukemic cells.Fresh leukemic cells from 25 AML, 10 ALL, and 1 acute mixed-lineage leukemia (AMLL) patients were FACS-analyzed for the expression of CD34, CD33, CD38, and HLA-DR and examined for the expression levels of the WT1 gene (Table 2). WT1 expression levels of individual leukemic cell samples were plotted according to the levels of expression of CD34, CD33, CD38, and HLA-DR, which were either expressed or not expressed in the majority (>80%) of the leukemic cells (Fig 3). The average WT1 expression levels in leukemic cells were 2.4 to 9.3 × 10−1 and at least 10 times higher than those in hematopoietic progenitor cells of BM and CB, regardless of the expression of CD34, CD33, CD38, or HLA-DR. Next, fresh leukemic cells from 5 patients with AML were FACS-sorted according to the expression levels of CD34 and CD33 (Fig 1C). The average levels of WT1 expression in FACS-sorted CD34+CD33−, CD34+CD33+, CD34−CD33+, and CD34−CD33− leukemic cell populations from 5 AML samples were 9.3 × 10−1, 5.6 × 10−1, 5.5 × 10−1, and 4.5 × 10−1, respectively (Figs 2 and 3 and Table 1). All of the four sorted leukemic cell populations expressed high levels of WT1, reflecting the phenotypical heterogeneity of leukemic cells. Moreover, various leukemic cell lines were examined for cell surface markers and WT1 expression (Fig 3 and Table 2). The WT1 expression levels in all but four leukemic cell lines examined (Molt-3, Tall-1, Jurkat, and Ball-1) were at least 13 times higher than those in hematopoietic progenitor cells of BM and CB, regardless of the expression of CD34, CD33, CD38, HLA-DR, and c-kit. Finally, fresh leukemic cells were used to analyze whether WT1 expression levels are related to the expression of CD34, CD33, CD38, and HLA-DR. WT1 expression levels were found to be significantly higher in CD34+HLA-DR− than in CD34+HLA-DR+ leukemic cells (P < .001), and in CD34−CD33+ than in CD34−CD33− leukemic cells (P < .05).
Correlation Between WT1 Gene Expression Levels and Surface Markers
| Leukemic Samples . | ||||||
|---|---|---|---|---|---|---|
| FAB . | Patient No. . | WT1 mRNA . | CD34 . | CD33 . | CD38 . | HLA-DR . |
| L1 | 289 | 0.93 | + | − | + | − |
| M1 | 80 | 0.57 | + | − | − | + |
| AMLL | 51 | 0.44 | + | − | + | + |
| ALL | 81 | 0.34 | + | − | + | ND |
| ALL | 197 | 0.25 | + | − | − | + |
| M2 | 124 | 0.11 | + | − | ND | + |
| M2 | 237 | 0.074 | + | − | + | + |
| M2 | 220 | 0.022 | + | − | + | ND |
| L2 | 234 | 0.021 | + | − | + | ND |
| L1 | 275 | 0.012 | + | − | − | + |
| M2 | 273 | 0.89 | + | + | ND | − |
| M3 | 228 | 0.84 | + | + | − | − |
| L2 | 139 | 0.70 | + | + | ND | + |
| M1 | 6 | 0.45 | + | + | ND | + |
| M4 | 294 | 0.41 | + | + | + | + |
| MDS-AML | 318 | 0.30 | + | + | + | ND |
| MDS-AML | 243 | 0.20 | + | + | ND | + |
| M4 | 227 | 0.05 | + | + | + | ND |
| ALL | 288 | 0.026 | + | + | + | ND |
| M2 | 199 | 0.026 | + | + | + | + |
| M5a | 37 | 1.72 | − | + | ND | + |
| M1 | 317 | 1.34 | − | + | + | − |
| M3 | 282 | 1.01 | − | + | ND | − |
| M6 | 287 | 0.98 | − | + | + | + |
| M4 | 275 | 0.97 | − | + | + | + |
| M5a | 306 | 0.62 | − | + | + | + |
| M3 | 78 | 0.58 | − | + | ND | − |
| M2 | 235 | 0.45 | − | + | + | − |
| M5 | 296 | 0.11 | − | + | + | + |
| M3 | 137 | 0.025 | − | + | ND | − |
| M5 | 274 | 0.013 | − | + | + | + |
| M5b | 271 | 0.010 | − | + | ND | + |
| L2 | 5 | 0.40 | − | − | + | − |
| ALL | 280 | 0.25 | − | − | + | ND |
| M5a | 33 | 0.050 | − | − | + | + |
| L2 | 151 | 0.017 | − | − | + | + |
| Leukemic Samples . | ||||||
|---|---|---|---|---|---|---|
| FAB . | Patient No. . | WT1 mRNA . | CD34 . | CD33 . | CD38 . | HLA-DR . |
| L1 | 289 | 0.93 | + | − | + | − |
| M1 | 80 | 0.57 | + | − | − | + |
| AMLL | 51 | 0.44 | + | − | + | + |
| ALL | 81 | 0.34 | + | − | + | ND |
| ALL | 197 | 0.25 | + | − | − | + |
| M2 | 124 | 0.11 | + | − | ND | + |
| M2 | 237 | 0.074 | + | − | + | + |
| M2 | 220 | 0.022 | + | − | + | ND |
| L2 | 234 | 0.021 | + | − | + | ND |
| L1 | 275 | 0.012 | + | − | − | + |
| M2 | 273 | 0.89 | + | + | ND | − |
| M3 | 228 | 0.84 | + | + | − | − |
| L2 | 139 | 0.70 | + | + | ND | + |
| M1 | 6 | 0.45 | + | + | ND | + |
| M4 | 294 | 0.41 | + | + | + | + |
| MDS-AML | 318 | 0.30 | + | + | + | ND |
| MDS-AML | 243 | 0.20 | + | + | ND | + |
| M4 | 227 | 0.05 | + | + | + | ND |
| ALL | 288 | 0.026 | + | + | + | ND |
| M2 | 199 | 0.026 | + | + | + | + |
| M5a | 37 | 1.72 | − | + | ND | + |
| M1 | 317 | 1.34 | − | + | + | − |
| M3 | 282 | 1.01 | − | + | ND | − |
| M6 | 287 | 0.98 | − | + | + | + |
| M4 | 275 | 0.97 | − | + | + | + |
| M5a | 306 | 0.62 | − | + | + | + |
| M3 | 78 | 0.58 | − | + | ND | − |
| M2 | 235 | 0.45 | − | + | + | − |
| M5 | 296 | 0.11 | − | + | + | + |
| M3 | 137 | 0.025 | − | + | ND | − |
| M5 | 274 | 0.013 | − | + | + | + |
| M5b | 271 | 0.010 | − | + | ND | + |
| L2 | 5 | 0.40 | − | − | + | − |
| ALL | 280 | 0.25 | − | − | + | ND |
| M5a | 33 | 0.050 | − | − | + | + |
| L2 | 151 | 0.017 | − | − | + | + |
| Leukemia and Lymphoma Cell Lines . | |||||||
|---|---|---|---|---|---|---|---|
| Cell Line . | Origin . | WT1 mRNA . | CD34 . | CD33 . | CD38 . | HLA-DR . | c-kit . |
| Molt-4 | T-ALL | 0.41 | + | − | + | − | − |
| KG-1 | M1 | 1.12 | + | + | − | − | +low |
| KU812 | CML | 0.83 | + | + | + | − | +low |
| TF-1 | M6 | 0.80 | + | + | + | + | +low |
| Kasumi-1 | M2 | 0.60 | + | + | + | + | +low |
| BV173 | CML | 1.50 | − | + | + | + | − |
| K562 | CML | 1.00 | − | + | − | − | − |
| HL-60 | M3 | 0.56 | − | + | + | − | − |
| THP-1 | M5 | 0.45 | − | + | + | − | − |
| HEL | M6 | 0.37 | − | + | − | + | +low |
| IMS-M1 | M5 | 0.31 | − | + | + | − | − |
| SAM-1 | CML | 0.71 | − | − | − | − | − |
| Molt-3 | T-ALL | 0.10 | − | − | + | − | − |
| Tall-1 | T-ALL | 0.04 | − | − | + | − | − |
| Jurkat | T-ALL | 2.0 × 10−4 | − | − | + | − | − |
| Ball-1 | B-ALL | <10−5 | − | − | + | + | ND |
| Daudi | Lymphoma | 3.8 × 10−4 | − | − | + | + | − |
| U937 | Lymphoma | <10−5 | − | + | + | − | − |
| KT-3 | Lymphoma | <10−5 | − | − | − | + | − |
| DL-40 | Lymphoma | <10−5 | − | − | + | + | − |
| Leukemia and Lymphoma Cell Lines . | |||||||
|---|---|---|---|---|---|---|---|
| Cell Line . | Origin . | WT1 mRNA . | CD34 . | CD33 . | CD38 . | HLA-DR . | c-kit . |
| Molt-4 | T-ALL | 0.41 | + | − | + | − | − |
| KG-1 | M1 | 1.12 | + | + | − | − | +low |
| KU812 | CML | 0.83 | + | + | + | − | +low |
| TF-1 | M6 | 0.80 | + | + | + | + | +low |
| Kasumi-1 | M2 | 0.60 | + | + | + | + | +low |
| BV173 | CML | 1.50 | − | + | + | + | − |
| K562 | CML | 1.00 | − | + | − | − | − |
| HL-60 | M3 | 0.56 | − | + | + | − | − |
| THP-1 | M5 | 0.45 | − | + | + | − | − |
| HEL | M6 | 0.37 | − | + | − | + | +low |
| IMS-M1 | M5 | 0.31 | − | + | + | − | − |
| SAM-1 | CML | 0.71 | − | − | − | − | − |
| Molt-3 | T-ALL | 0.10 | − | − | + | − | − |
| Tall-1 | T-ALL | 0.04 | − | − | + | − | − |
| Jurkat | T-ALL | 2.0 × 10−4 | − | − | + | − | − |
| Ball-1 | B-ALL | <10−5 | − | − | + | + | ND |
| Daudi | Lymphoma | 3.8 × 10−4 | − | − | + | + | − |
| U937 | Lymphoma | <10−5 | − | + | + | − | − |
| KT-3 | Lymphoma | <10−5 | − | − | − | + | − |
| DL-40 | Lymphoma | <10−5 | − | − | + | + | − |
Abbreviation: ND, not determined; FAB, French-American-British criteria.
DISCUSSION
The WT1 expression in both the most primitive cells (CD34+CD33−,11-17 CD34+CD38−,18-20 CD34+HLA-DR−,21-27 or CD34+c-kitlow 9,28-35) and the more mature progenitor cells (CD34+CD33+, CD34+CD38+, CD34+HLA-DR+, or CD34+c-kithigh) of BM and CB was identified at very low (1.0 to 2.4 × 10−2) or undetectable (<10−2) levels. Moreover, the WT1 expression in more differentiated CD34− cells was undetectable in all of the 5 CD34− cell samples. However, these results cannot exclude the possibility of a transient expression of WT1 among a subpopulation of hematopoietic precursors. On the other hand, fresh leukemic cells expressed high levels (average, 2.4 to 9.3 × 10−1) of WT1, regardless of cell surface markers. Thus, the WT1 expression levels in normal hematopoietic progenitor cells were at least one log less than those in leukemic cells. Before we obtained the present data, we assumed that we could easily find normal counterparts for the leukemic cells with comparable WT1 expression levels; however, we could not. This suggests that overexpression of the WT1 gene in leukemic cells reflects a leukemic state of hematopoietic progenitor cells, but not WT1 expression levels in normal counterparts.
Fraizer et al36 have recently reported that CD34+ BM cells express WT1 at levels comparable to those in K562 and fresh leukemic cells. Their results are not compatible with ours in that the expression levels of WT1 not only in the CD34+and CD34− cell populations, but also in the CD34+CD33−, CD34+CD33+, CD34+CD38−, CD34+CD38+, CD34+HLA-DR−, CD34+HLA-DR+, CD34+c-kithigh, CD34+c-kitlow, and CD34+c-kit− subsets were very low (1.0 to 2.4 × 10−2) or undetectable (<10−2) and at least 10 times less than those in leukemic cells. The precise cause of this discrepancy between their results and ours is unknown. However, it might be ascribed to the difference in the optimal conditions for RT-PCR (our RT-PCR conditions were optimized as described here and previously5,6 ). Menssen et al37 used an immunofluorescence assay using the anti-WT1 MoAb and found the WT1 protein only in the nuclei of leukemic blast cells, but not in the nuclei of normal CD34+ hematopoietic progenitor cells. Moreover, the WT1 transcript was detected by RT-PCR assay in 68 of 86 (79%) ALL patients and in 53 of 57 (93%) AML patients. In contrast, normal peripheral CD34+ hematopoietic progenitor cells did not express the WT1 gene at detectable levels. Thus, the data presented here are basically compatible with the findings of Menssen et al, although the detection limit (1 in 5,000) for WT1 transcript of their RT-PCR assay is 20 times lower than that (1 in 100,000) of our RT-PCR assay, and it remains unclear whether peripheral CD34+ cells can be compared with the CD34+ cells of BM and CB.
Of 5 CD34+ cell samples, 4 expressed very low levels (1.0 to 2.4 × 10−2) of WT1, whereas the WT1 expression levels in all of the 5 CD34− cell samples were undetectable (<10−2). Thus, the WT1 expression levels were significantly (P < .01) higher in CD34+ cells than in CD34− cells, suggesting that the WT1 expression levels decrease as hematopoietic progenitor cells differentiate into more mature cells. Moreover, the WT1 expression levels in the CD34+HLA-DR− and CD34−CD33+ leukemic cells were respectively and significantly higher than those in the more differentiated CD34+HLA-DR+ and CD34−CD33− leukemic cells. The WT1 expression levels in mature T- and B-cell ALL cell lines (Molt-3, Tall-1, Jurkat, and Ball-1) were very low. This is consistent with the findings that the WT1 expression was downregulated during the differentiation of HL60 cells by dimethyl sulfoxide or retinoic acid38 and during induction of erythroid and megakaryocytic differentiation of K562 cells.39
When leukemic cell samples from 15 acute leukemia patients were subjected to PCR single-strand conformation polymorphism (SSCP) analysis, no point mutations were found within the zinc finger region.5 Furthermore, when 12 of 15 leukemia samples were examined by SSCP analysis for point mutations within the region (exons 2 to 6) 5′ to the zinc finger region, no point mutations were detected.5 On the other hand, King-Underwood et al40 have recently reported that they have found mutations in the WT1 gene in 4 of 36 acute leukemia samples by SSCP analysis. The mutations comprise small insertions in exons 1 and 7 and a nonsense mutation in exon 9, predicting the production of a truncated WT1 protein with absence or disruption of the zinc finger region. Determination of biological significance of WT1 mutations should clarify the roles of the WT1 gene in leukemogenesis.
The mechanism of overexpression of the WT1 gene in human leukemic cells remains undetermined. Recently, Wu et al41 characterized the human WT1 enhancer region located at the 3′ of the WT1 gene, and showed that the activity of the 3′ WT1 enhancer was positively regulated by erythrocyte/megakaryocyte-related transcription factor GATA-1. Shimamoto et al42 reported that GATA-1 was expressed in 30 of 62 AML, 2 of 19 ALL, and 9 of 14 CML blastic crisis. Thus, GATA-1 expression might play a part in WT1 overexpression in leukemia. However, GATA-1 expression is limited to a part of leukemia, whereas WT1 overexpression is general in leukemia. Therefore, as yet undetermined mechanisms should play an important role in an aberrant overexpression of WT1 in leukemia.
Fresh leukemic cells were FACS-sorted and fractionized into four populations (CD34+CD33−, CD34+CD33+, CD34−CD33+, and CD34−CD33−) according to the expression levels of CD34 and CD33. Interestingly, all four populations expressed high levels of WT1, suggesting that these four populations all consisted of leukemic cells and also that these leukemic cells were phenotypically heterogenous. These results indicate that a leukemic clone that was generated by malignant transformation of a hematopoietic progenitor cell became heterogenous during its proliferation and expansion.
Mononuclear cells of normal BM expressed 10−4 levels of WT1.5 It remains unclear which cells are responsible for these background levels of WT1 expression, because the present study could not definitively identify such cells. However, because low levels of WT1 (10−2 levels) were expressed in 4 of the 5 CD34+ cell samples and in 17 of the 55 CD34+ subsets, whereas in all of the 5 CD34− cell samples WT1 expression was at undetectable levels (<10−2), CD34+ cells appear to be responsible for the background levels of WT1 expression in BM.
ACKNOWLEDGMENT
We thank Tsuyomi Noguchi and Hiromi Takeuchi for typing this manuscript and Machiko Mishima for her skillful technical assistance with the PCR.
Address reprint requests to Haruo Sugiyama, MD, Departments of Clinical Laboratory Science and Medicine III, Osaka University Medical School, 2-2, Yamada-Oka, Suita City, Osaka 565, Japan.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal