Abstract
The integrin α2β1 is a receptor for collagen that plays a fundamental role in the adhesion of blood platelets to the extracellular matrix. We previously reported that platelet α2β1 levels among randomly selected individuals can vary up to 10-fold and that this correlates with differences in adhesiveness to type-I or type-III collagens. We have now found two linked, allelic polymorphisms within the coding sequence of the α2 gene that correlate with receptor density, TTT/TTC at codon Phe224 and ACA/ACG at codon Thr246. By Southern blot hybridization of specific antisense DNA probes to segments of genomic DNA that encompass each coding region, we have determined the gene frequencies of each allele in a random donor population (n = 65) to be 0.585 (TTC ... ACG) and 0.415 (TTT ... ACA). There is a statistically significant correlation between the alleles TTT ... ACA (codons 224…246) and high receptor density (n = 30; P < .002), whereas the complimentary alleles TTC ... ACG are associated with low receptor density. Heterozygous individuals express intermediate levels of this receptor, and familial studies confirm that these allelic polymorphisms are inherited characteristics. These findings prove that the level of platelet α2β1 is an inherited trait. The molecular basis for receptor density remains to be determined, but our findings establish that these silent alleles within the coding sequence of the α2 gene are linked to the genetic basis for variation in receptor density.
THE INTEGRIN α2β1 MEDIATES platelet adhesion to both fibrillar (types I-III and V) or nonfibrillar (types IV, VI, VII, and VIII) collagens.1-6 The extent to which this receptor contributes to adhesion to any one type of collagen is affected by both divalent cation composition and fluid shear rate.6 In whole blood, α2β1 -mediated adhesion of platelets to fibrillar collagens, including types I and III, also activates platelets, leading to prothrombin conversion on the platelet surface7 and thrombus formation mediated by another integrin, αIIbβ3 .8,9 However, even in the presence of inhibitors of αIIbβ3 function, platelet activation initiated by α2β1 -dependent adhesion leads to cellular events that catalyze prothrombin conversion and fibrin clot formation.7 Thus, α2β1 has the potential to contribute significantly to platelet function in vivo. Indeed, patients with quantitative abnormalities of platelet α2 present with prolonged bleeding times, chronic mucocutaneous bleeding, defective in vitro platelet adhesion to collagen and absent in vitro collagen-induced aggregation.10-12 Moreover, selected individuals with autoimmune platelet dysfunction have been found to express serum autoantibodies specific for α2 that block in vitro platelet adhesion to collagen and collagen-induced aggregation.13 14
Although the levels of the integrins α5β1 or αIIbβ3 and the glycoprotein Ib-IX complex on platelets (molecules per platelet) can vary from one individual to the next,15,16 these differences never exceed a fraction of the mean population level. On the other hand, the number of α2β1 molecules per platelet varies by as much as an order of magnitude and correlates precisely with quantitative estimates of platelet adhesion to type-I or type-III collagens.15 In this report, we provide direct evidence for a genetic basis for the observed difference in platelet α2β1 density.
MATERIALS AND METHODS
Reagents.Murine monoclonal IgG antibody 6F1 (anti-α2β1 ) has been described8 and is a gift from Dr B. Coller (Mount Sinai Medical Center, New York, NY). The murine hybridoma 12F1 producing IgG specific for α2β1 has been well characterized17 and was generously provided by Dr V. Woods (University of California at San Diego, La Jolla, CA). The murine monoclonal IgG antibody 8C12, also specific for the α2β1 , was a gift from Dr M. Ginsberg (The Scripps Research Institute, La Jolla, CA). AP2 is a murine monoclonal IgG antibody specific for the integrin αIIbβ3 developed and characterized in our laboratory.18
mRNA amplification and sequencing.Total platelet RNA was isolated as described,19 and α2 -specific mRNA was amplified as four overlapping segments by reverse transcription-polymerase chain reaction (RT-PCR)20 using either Taq I (Perkin Elmer, Foster City, CA) or VENT (New England Biolabs, Beverly, MA) polymerases. The oligonucleotide primer 5′-CACTTAAGGCTTGGAAACTG-3′ representing bp 2772-2791 of mature α2 mRNA21 was used for the production of first-strand cDNA using RT. Two-stage PCR reactions were then used to amplify each of four cDNA segments, designated A through D, from 5′ to 3′. These oligonucleotide primers were as follows: for segment A, GAATTCCTGCAAACCCAGCG (bp 1-20) and CTGTTTAAGTACCCAAGAACTGC (bp 976-998) followed by ATTCtcGagAACCCAGCGCAACTACGG (bp 12-29)and GTTTctagACCCAAGAACTGCTATGCC (bp 970-988); for segment B, GAATTCCTGCAAACCCAGCG (bp 1-20) and CACTTAAGGCTTGGAAACTG (bp 2772-2791) followed by GCActcgagGGTGGGCGACGAAGTGCTACG (bp853-873) and TCCtctaGATCCCAAGATTTTCTGGG (bp 1850-1868); for segment C, GGTTCACCACTAGAAAATCAG (bp 1771-1791) and CACTTAAGGCTTGGAAACTG (bp 2772-2791) followed by TCACtcgagGAAAATCAGAATTCTGGAGC (bp 1783-1802) and ACTTctaGaTTGGAAACTGAGAGACGCC (bp 2763-2781); and for segment D, CAGCAGCTTCTGGTGGGCGAC(bp 842-862) and GTCTGCTGGTTCAGCTACTGAGC (bp 3582-3604) followed by GCTctcgagCAGAAGTCTGTTGCCTGCG (bp 2659-2677) and GCTtcTagAGCTACTGAGCTCTGTGG (bp 3575-3592). In each case, the Xho I or Xba I restriction sites used for cloning into M13 are underlined, and lowercase letters indicate base changes introduced to create these restriction sites. For reference, the start codon begins at base 49, whereas the stop codon begins at base 3592.21 cDNA segments were sequenced by the dideoxy termination method using Sequenase 2.0 (Promega Biotec, Inc, Madison, WI).
Southern blot hybridization.These studies were designed to identify the specific allelic polymorphisms at codons 807 and 873 of the α2 coding sequence. First-strand cDNA was generated using RT as described above. The product served as a template for a primary PCR using primers 5′-GAATTCCTGCAAACCCAGCG-3′ (bp 1-20) and 5′-CTGTTTAAGTACCCAAGAACTGC-3′ (bp 976-998). The desired segment of this product was further amplified in a secondary PCR using the primer pair ATTCtcGagAACCCAGCGCAACTACGG and GTTTctagACCCAAGAACTGCTATGCC to produce a segment corresponding to base pairs 12-988 or using the primer pair GAACTCGAGGTTTTCTCACATGTGG and CTGTTTAAGTACCCAAGAACTGC to produce a segment corresponding to base pairs 419-998. Vent DNA polymerase (New England Biolabs, Inc, Beverly, MA) was used in all PCR reactions.
The desired segment of the α2 gene was also amplified using leukocyte DNA as the template. PCR was performed using primer pair GTGTTTAACTTGAACACATATAAAACC (bp 715-741) plus CTCAGTATATTGTCATGGTTGCATTG (bp 940-965) in the first stage and then the nested primer pair TCCCtcgAgTCCCAATATGGTGGGGACCTCAC (Xho I: bp 775-797) plus CTCtcTAgATTGTCATGGCATTGATCAATCAC (bp 931-956: Xba I) in the second stage. The product is an ≈4.5-kb DNA segment that includes both the 807 and 873 allelic sequences and 4.35 kb of intervening, noncoding sequence. It is likely that this represents a single large intron separating two exons that encode each of the allelic sequences. The Xho I and Xba I restriction sites were introduced to facilitate subcloning of the segment. On digestion with Xba I, it became apparent that there was one additional restriction site within the noncoding sequence. Thus, complete digestion of cloned DNA with Xho I and Xba I yields a 3.7-kb 5′ fragment and a 650-bp 3′ fragment. The sequence of the entire 650-bp fragment as well as over 600 bp of the larger 3.7-kb fragment were obtained by the dideoxy method using Sequenase 2.0 (Promega Biotec, Inc).
Using the newly found intron sequence, PCR conditions were established to amplify smaller genomic DNA fragments that would include either the 807 or 873 allelic sequences, so that these could be used in Southern blot hybridization assays. A 298-bp genomic fragment that includes the 807 alleles is amplified with the primer pair GTGTTTAACTTGAACACATATAAAACC (bp 715-741) and GATTTAACTTTCCCAGCTGCCTTC (intron sequence 173-196). The allelic sequences at 807 are then detected by hybridization to the probes 807C or 807T (Table 1). An oligonucleotide corresponding to intron sequence ACATTGGCCTATTAGCA serves as a positive control. At the same time, a 398-bp genomic fragment that includes the 873 alleles is amplified with primer pair CGAATACTGGGATAAATACATGCAC (intron sequence) and CTCAGTATATTGTCATGGTTGCATTG (bp 940-965). The allelic sequences at 873 are then detected by hybridization to the probes 873G or 873A (Table 1). An oligonucleotide corresponding to coding sequence 887-903 serves as the positive control for this DNA segment.
Southern Blot Oligonucleotide Probes
| Name . | Sequence . | bp* . |
|---|---|---|
| 807C | ATTGCTCCGAATGTGTT | 799-815 |
| 807T | ATTGCTCCAAATGTGTT | 799-815 |
| Intron | ACATTGGCCTATTAGCA | |
| 873G | ATTACTTTCGTAGCACT | 865-881 |
| 873A | ATTACTTTTGTAGCACT | 865-881 |
| Exon | TTCACCGTCAGTTACAA | 887-903 |
| Name . | Sequence . | bp* . |
|---|---|---|
| 807C | ATTGCTCCGAATGTGTT | 799-815 |
| 807T | ATTGCTCCAAATGTGTT | 799-815 |
| Intron | ACATTGGCCTATTAGCA | |
| 873G | ATTACTTTCGTAGCACT | 865-881 |
| 873A | ATTACTTTTGTAGCACT | 865-881 |
| Exon | TTCACCGTCAGTTACAA | 887-903 |
For each set of allelic probes, the distinctive base is indicated in boldface.
Sequence numbering from Takada and Hemler.21
DNA products were denatured with 0.36 mol/L NaOH plus 25 mmol/L EDTA, followed by neutralization with 1 mol/L ammonium acetate (final concentration). The DNA samples were applied to a prewetted nylon membrane using a vacuum dot-blot apparatus, and the DNA was fixed to the membrane by cross-linking with UV light. The blots were prehybridized by incubation in 5× SSC (saline sodium citrate), 0.2% sodium dodecyl sulfate, 0.1% hybridization component (Amersham, Arlington Heights, IL), plus 0.5% blocking agent (Amersham) for 3 hours. Fluorescein-labeled oligonucleotide probes were then added at a final concentration of 5 ng/mL. After incubation at 39°C for 12 to 16 hours, each blot was subjected to specific stringency washes. We used two oligonucleotide probes to detect each of the two alleles of codons 807 and 873 (Table 1). Each oligonucleotide probe was labeled using the enhanced chemiluminescence 3′-oligolabeling and detection system from Amersham Life Science, Inc, according to the manufacturer's instructions. This method uses terminal transferase to conjugate the 3′ end of the probe with fluorescein-deoxyuridine triphosphate. Bound probe is detected with horseradish peroxidase-conjugated, antifluorescein antibody and subsequent addition of enhanced chemiluminescence detection reagents supplied with the Amersham kit.
Quantitation of platelet α2β1. Murine monoclonal antibodies specific for the α2β1 complex, 6F1, 12F1, and 8C12, were used to quantitate levels of this receptor on platelets by flow cytometry or direct binding of radioiodinated Fab molecules, as previously described.15 For analyses of washed platelets by flow cytometry (as in family study no. 2), bound murine antibodies 6F1 or AP2 (anti-αIIbβ3 ) were detected using a 1:50 dilution of fluorescein isothiocyanate (FITC)-F(ab′)2 goat antimouse IgG (heavy and light chains; Zymed Laboratories, Inc, South San Francisco, CA). Levels of bound 6F1 or AP2 were then expressed by mean fluorescence intensity (MFI) after subtraction of MFI observed for nonimmune murine IgG. For flow cytometric assays of whole blood samples, platelets were gated by forward versus side scatter, and the levels of bound 12F1, 8C12, AP2, or nonimmune murine IgG (mIgGl) were determined using a single FITC-F(ab′)2 goat antimouse IgG (heavy and light chains; Zymed). The levels of bound 12F1 and 8C12 were first corrected by subtracting the level of bound mIgG and were then normalized by dividing by the corrected MFI obtained with AP2, using the formulas: n8C12 = 100 × [8C12 − mIgG] ÷ [AP2 − mIgG] or n12F1 = 100 × [12F1 − mIgG] ÷ [AP2 − mIgG].
Platelet adhesion to human collagens.Platelet adhesion to purified type-I or type-III human collagens was quantitated exactly as descibed.15
RESULTS
Based on the results from six individuals, we found no differences in the deduced amino acid sequence of α2 relative to the published sequence of Takada and Hemler,21 except for the allelic Br polymorphism at codon 505 (Lys/Glu).22 On the other hand, we noted two conservative differences within the coding sequence (C/T at bp 807 and G/A at bp 873) that apparently are allelic polymorphisms.
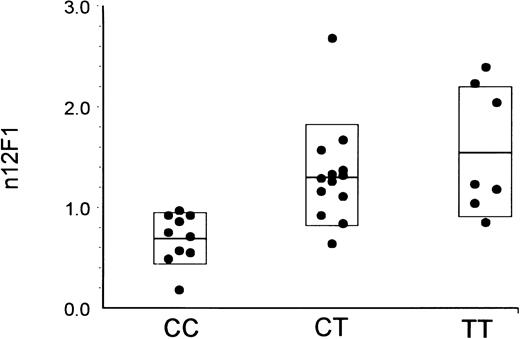
We sought to determine whether there is a correlation between these α2 sequence differences and the level of the integrin α2β1 on platelets of a panel of normal subjects. Platelet α2β1 levels of 30 individuals were measured in whole blood samples by flow cytometry. In Fig 1, individual platelet α2β1 levels, as determined by the binding of monoclonal antibody 12F1, are plotted according to the α2 genotype of each individual, as determined by hybridization of specific allelic probes to genomic DNA. There is an obvious association of 807C and 873G with low receptor densities; conversely, increasing receptor density is associated with the alleles 807T and 873A. Equivalent findings were made with antibody 8C12 (data not shown). A 1-way analysis of variance using the Student-Newman-Keuls method indicates that there is a statistically significant difference between genotype and α2β1 level (for 8C12, P = .000188; for 12F1, P = .00147).
Relationship between α2 807 genotype and platelet α2β1 levels. The level of platelet α2β1 was determined by flow cytometry using the binding of monoclonal antibody 12F1. The corrected and normalized MFI values are indicated on the ordinate, and the results from each donor are plotted as a function of donor genotype with respect to the 807C and 807T alleles. Values are single measurements from each of 30 individuals but are representative of repeated measurements performed on several occasions. The boxed area in each data set represents the mean ± 1 SD. The difference in the means between donor groups is statistically significant (CC v CT, P < .05; and CC v TT, P < .05).
Relationship between α2 807 genotype and platelet α2β1 levels. The level of platelet α2β1 was determined by flow cytometry using the binding of monoclonal antibody 12F1. The corrected and normalized MFI values are indicated on the ordinate, and the results from each donor are plotted as a function of donor genotype with respect to the 807C and 807T alleles. Values are single measurements from each of 30 individuals but are representative of repeated measurements performed on several occasions. The boxed area in each data set represents the mean ± 1 SD. The difference in the means between donor groups is statistically significant (CC v CT, P < .05; and CC v TT, P < .05).
Allele frequencies were then calculated from a larger group of 65 randomly selected individuals. Based on these results, the gene frequencies for the 807C (or 873G) and 807T (or 873A) allelic pairs are 0.585 and 0.415, respectively. Furthermore, this extended comparison of the 807 and 873 alleles confirms linkage between the two polymorphisms.
We next analyzed the inheritance of collagen receptor density and these allelic sequences in the immediate family of donor no. 1, who was studied in our previous report15 and who expresses the lowest platelet receptor density of all donors examined to date. The platelet α2β1 density of each child (donors no. 16 and 17), as measured by the direct binding of antibody 125I-6F1 Fab fragments to washed platelets, is almost precisely the arithmetic mean of the platelet receptor densities of the parents (Table 2). The genotype of each family member was determined by Southern blot analysis of cDNA. Donor no. 1 (low receptor density) expresses only the 807C allele, whereas donor no. 15 (high receptor density) expresses only the 807T allele. The two offspring (donors no. 16 and 17) are obligate heterozygotes and express both the 807C and 807T alleles. The control oligonucleotide 887 bound to all cDNA samples, as expected. Each cDNA sample gave an identical result when probes specific for the alleles 873G and 873A were used (data not shown).
Platelet Integrin α2β1
| Family No. 1 . | 807 Alleles . | Antigen (Radioimmunoassay)* . | Activity† . | |||
|---|---|---|---|---|---|---|
| . | . | AP2 (anti-αIIbβ3) . | 6F1 (anti-α2β1) . | Type-I Collagen . | Type-III Collagen . | Fibronectin . |
| Mother (no. 1) | CC | 52,714 ± 442 | 933 ± 34 | 0.7 ± 0.2 | 3.5 ± 0.2 | 11.1 ± 1.2 |
| Father (no. 15) | TT | 49,102 ± 547 | 2,325 ± 122 | 16.8 ± 1.2 | 18.7 ± 0.6 | 9.1 ± 0.6 |
| Son (no. 16) | CT | 49,613 ± 877 | 1,612 ± 213 | 6.1 ± 0.7 | 8.7 ± 1.3 | 10.0 ± 1.2 |
| Son (no. 17) | CT | 49,311 ± 342 | 1,606 ± 127 | 2.2 ± 0.6 | 4.3 ± 1.0 | 8.9 ± 0.8 |
| Family No. 2 | 807 Alleles | Antigen (Flow Cytometry)‡ | Activity† | |||
| AP2 (anti-αIIbβ3) | 6F1 (anti-α2β1) | Type-I Collagen | Type-III Collagen | Fibronectin | ||
| Family No. 1 . | 807 Alleles . | Antigen (Radioimmunoassay)* . | Activity† . | |||
|---|---|---|---|---|---|---|
| . | . | AP2 (anti-αIIbβ3) . | 6F1 (anti-α2β1) . | Type-I Collagen . | Type-III Collagen . | Fibronectin . |
| Mother (no. 1) | CC | 52,714 ± 442 | 933 ± 34 | 0.7 ± 0.2 | 3.5 ± 0.2 | 11.1 ± 1.2 |
| Father (no. 15) | TT | 49,102 ± 547 | 2,325 ± 122 | 16.8 ± 1.2 | 18.7 ± 0.6 | 9.1 ± 0.6 |
| Son (no. 16) | CT | 49,613 ± 877 | 1,612 ± 213 | 6.1 ± 0.7 | 8.7 ± 1.3 | 10.0 ± 1.2 |
| Son (no. 17) | CT | 49,311 ± 342 | 1,606 ± 127 | 2.2 ± 0.6 | 4.3 ± 1.0 | 8.9 ± 0.8 |
| Family No. 2 | 807 Alleles | Antigen (Flow Cytometry)‡ | Activity† | |||
| AP2 (anti-αIIbβ3) | 6F1 (anti-α2β1) | Type-I Collagen | Type-III Collagen | Fibronectin | ||
| Mother (no. 18) | CC | 49.35 | 1.28 | 5.5 ± 0.4 | 12.1 ± 0.4 | 13.5 ± 3.2 |
| Father (no. 19) | CT | 51.01 | 4.15 | 17.5 ± 1.1 | 29.9 ± 1.3 | 14.0 ± 1.3 |
| Son (no. 20) | CT | 48.11 | 3.54 | 17.9 ± 1.2 | 25.2 ± 1.1 | 9.9 ± 2.9 |
| Daughter (no. 21) | CC | 44.91 | 1.42 | 3.4 ± 0.9 | 11.2 ± 1.0 | 10.1 ± 1.0 |
| Mother (no. 18) | CC | 49.35 | 1.28 | 5.5 ± 0.4 | 12.1 ± 0.4 | 13.5 ± 3.2 |
| Father (no. 19) | CT | 51.01 | 4.15 | 17.5 ± 1.1 | 29.9 ± 1.3 | 14.0 ± 1.3 |
| Son (no. 20) | CT | 48.11 | 3.54 | 17.9 ± 1.2 | 25.2 ± 1.1 | 9.9 ± 2.9 |
| Daughter (no. 21) | CC | 44.91 | 1.42 | 3.4 ± 0.9 | 11.2 ± 1.0 | 10.1 ± 1.0 |
The values shown are the number of molecules bound per platelet (mean ± SD; n = 3).
The values shown are the number of platelets × 10−5 bound as indicated (mean ± SD; n = 3 for family no. 1 and n = 2 for family no. 2).
The values shown are MFIs.
In a second family study, flow cytometry was used to quantitate receptor densities on washed platelets (donors no. 18, 19, 20, and 21; see Table 2). AP2 was used to quantitate αIIbβ3 , whereas 6F1 was used to quantitate α2β1 . Bound primary antibody was detected with FITC-conjugated goat antimouse IgG. Comparisons of MFI give a valid estimate of receptor levels among family members. The genotype of individuals in family no. 2 was determined by hybridization of labeled probes to amplified segments of genomic DNA. The mother (donor no. 18) in this family is homozygous for the 807C allele, whereas the father (donor no. 19) is heterozygous. A son (donor no. 20), whose antigen level mirrors that of the father, is also heterozygous, whereas a daughter (donor no. 21), whose antigen level is similar to that of the mother, is also homozygous for the 807C allele. Comparable findings were made with probes specific for the 873 alleles (not shown).
DISCUSSION
From a comparison of the sequences of platelet α2 cDNA derived from several individuals, we were able to identify allelic polymorphisms that are linked to the genetic basis of variation in α2β1 receptor density. These polymorphisms in codons Phe224 (TTT/TTC) and Thr246 (AGA/AGC) are conservative and do not alter the deduced amino acid sequence of the translated protein. However, the inheritance of these silent polymorphisms correlates precisely with the inheritance of receptor density. Our finding that these allelic differences are inherited strongly supports our hypothesis that differences within the α2 gene are responsible for the variation in platelet α2β1 density that we previously reported.15
There are a number of possible explanations for our findings. On the one hand, the silent alleles within the coding sequence may be linked to other causative polymorphisms within the α2 gene, eg, variants of the α2 gene promoter region. The linkage of genetic mutations with otherwise silent polymorphisms within or outside of the coding sequence has been established in a number of inherited disorders, including pyruvate kinase deficiency23,24 and Gaucher disease.25,26 Alternatively, seemingly minor polymorphisms in mRNA sequence such as these may influence mRNA stability and/or turnover, resulting in disproportionate levels of each message.27 At this time, our results do not favor any one putative mechanism.
Even though α2β1 is widely distributed in a large variety of tissues, among the hematopoietic cells, it is constitutively expressed exclusively on megakaryocytes and platelets.28 Consistent with this observation, the megakaryoblastic cell line Dami constitutively expresses α2β1 ,29 and the level of α2β1 expressed by the erythroleukemic cell line K562 increases with the acquisition of the megakaryocytic phenotype induced by phorbol ester.30 This results from increased steady-state levels of α2 mRNA caused by transcriptional activation of the α2 gene.31 In contrast to these findings on megakaryocytic cells, T lymphocyte or monocyte expression requires sustained cell activation.32 The structural organization of a reported 5-kb promoter region of the α2 gene29,33 is very analogous to that of other megakaryocyte-specific genes, including PF4 and αIIb ,34 and several megakaryocyte-associated gene elements, such as GATA-like sequences and consensus binding sites for SAR/MAR proteins, are found in the most 5′ portion of this promoter region.29
Even though the extent of the variation in receptor density suggests that more than a single genetic factor is involved in establishing the number of α2β1 molecules per platelet in any one individual, our preliminary findings indicate that a significant correlation exists between receptor density and the cluster of linked polymorphisms that we have defined. Additional polymorphisms in either the coding sequence or noncoding sequence may well contribute to secondary variation in receptor density, but we maintain that we have already identified a polymorphic region of the gene that most likely correlates with differences in expression. It remains to be determined whether this region is itself responsible for the differences in receptor density or whether it is in linkage with another region of the gene that controls either transcription rate or stability/turnover of mRNA. Regardless of the outcome, our preliminary findings lay a solid foundation on which to investigate the genetic basis for the observed variation in α2β1 density among normal individuals.
The finding of an inherited basis for platelet α2β1 density has important implications in the diagnosis and treatment of cardiovascular disease. A number of hereditary and environmental factors most likely act in synergy to determine the overall risk for thrombosis and cardiovascular disease in any individual. Because blood platelets are responsible for the initiation of a thrombus, any variation in an adhesion receptor on blood platelets would logically become a potential risk factor. We postulate that increased expression of α2β1 can increase long-term risk of thrombotic disease, whereas decreased expression can impair hemostasis. It is not likely that heterogeneity of α2β1 alone compromises platelet function in an otherwise healthy individual, but inherited differences in receptor density could have an impact on the clinical outcomes in individuals who are at increased risk of thrombosis or bleeding as a result of unrelated disease processes or surgical interventions.
The potential impact of variation in α2β1 numbers on platelets must be given consideration because of the fact that fibrillar collagens are major components of the subendothelial matrix that can trigger thrombus formation in flowing blood. Platelets activated by contact with collagens can both participate in the formation of aggregates via fibrinogen binding to the integrin αIIbβ3 and provide sites for the assembly of coagulation factor VIIIa/IXa and factor Va/Xa complexes.35-39
ACKNOWLEDGMENT
We are especially grateful to Ms K. Cox and the General Clinical Research Center of the Scripps Clinic and Research Foundation for measurements of platelet receptor levels in whole blood samples by flow cytometry and isolation of mononuclear cells. We thank Mr R. Orchekowski and Dr Y. Honda for their valuable technical contributions during the initiation of these studies; Dr J. Ware (Scripps Research Institute) for his thoughtful contributions to our work and for sharing important protocols with us; and Dr E. Beutler (The Scripps Research Institute) for providing anonymous donor DNA samples. Monoclonal antibodies were provided by colleagues, and for these we wish to thank Dr B. Coller for monoclonal antibody 6F1, Dr M. Ginsberg (The Scripps Research Institute) for monoclonal antibody 8C12, and Dr V. Woods for the hybridoma cell line 12F1.
Supported by the National Heart, Lung, and Blood Institute Grant No. HL-46979 and by a grant from the Gustavus and Louise Pfeiffer Research Foundation (Redlands, CA). This is manuscript no. 8978-MEM from The Scripps Research Institute.
Address reprint requests to Thomas J. Kunicki, PhD, Department of Molecular and Experimental Medicine, The Scripps Research Institute, Room SR11, Maildrop SBR13, 10550 N Torrey Pines Rd, La Jolla, CA 92037.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal