Abstract
Severe combined immunodeficiency (SCID) is a syndrome of profoundly impaired cellular and humoral immunity. In humans, SCID is most commonly caused by mutations in the X-linked gene IL2RG, which encodes the common γ chain, γc, of the leukocyte receptors for interleukin-2 and multiple other cytokines. To investigate the frequency and variety of IL2RG mutations that cause SCID, we analyzed DNA, RNA, and B-cell lines from a total of 103 unrelated SCID-affected males and their relatives using a combination of molecular and immunologic techniques. Sixty-two different mutations spanning all eight IL2RG exons were found in 87 cases, making possible correlations between mutation type and functional consequences. Although skewed maternal X chromosome inactivation, single-strand conformation polymorphism, mRNA expression, and cell surface staining with anti-γc antibodies were all helpful in establishing IL2RG defects as the cause of SCID, only dideoxy fingerprinting and DNA sequence determination each detected 100% of the IL2RG mutations in our series. Abnormal γc chains may be expressed in the lymphocytes of as many as two thirds of patients with X-linked SCID. Specific mutation diagnosis thus remains technically challenging, but it is important for genetic counseling and perhaps for helping to select appropriate subjects for retroviral gene therapy trials.
SEVERE COMBINED immunodeficiency (SCID) is a rare syndrome of profoundly impaired cellular and humoral immune function.1-3 Without bone marrow transplantation (BMT), affected patients suffer severe and persistent infections, often with opportunistic pathogens, and generally die in infancy. Although both X-linked recessive and autosomal forms of SCID are recognized, the X-linked form is the most frequent. Patients with X-linked SCID generally have very low numbers of T cells and natural killer (NK) cells, whereas B cells are often found in relatively high numbers even though specific antibody responses are deficient.4 X-linked SCID is caused by mutations of IL2RG,5,6 the gene encoding the common γ chain, known as γc, found in the interleukin-2 (IL-2) receptor and multiple other cytokine receptors, including those for IL-4, IL-7, IL-9, and IL-15. The intracellular portion of γc is known to interact with Janus kinase 3 (Jak3), a signaling kinase that cooperates with other Jak and STAT proteins in a complex signal transduction array.7 8
Identification of IL2RG as the disease gene for X-linked SCID has made possible specific mutation diagnosis in individual patients with SCID. More than three dozen patient mutations have been reported in the literature by our group6,9-11 and by others,5,7 12-20 almost all of which have consisted of minor DNA sequence alterations. In this report, we present a total of 87 mutations in IL2RG that have caused SCID, a number large enough to give a new perspective on the mutational range of this gene both for diagnostic and clinical purposes and as a series of probes for studying the interaction of γc with its extracellular and intracellular contacts.
MATERIALS AND METHODS
Subjects and sample preparation.Males with SCID by criteria of the World Health Organization Working Group on immunodeficiencies3 were ascertained through immunologists and geneticists. Although those with adenosine deaminase deficiency (ADA-deficient SCID) or specific autosomal syndromes such as SCID with cartilage-hair hypoplasia were excluded, both pedigrees showing X-linked inheritance and sporadic cases were specifically sought. Clinical data and pedigrees were reviewed; all patients had T lymphocytopenia, although the majority had B cells in normal to elevated numbers. Relatedness among families in this study or between our patients and subjects of mutation reports by others was found retrospectively only in one instance.12 After receiving informed consent from the subjects, samples from the unrelated SCID-affected males and their relatives were obtained, including blood, fibroblast cell lines, fetal samples, and tissue from biopsy or autopsy. Where possible, B-cell lines transformed with Epstein-Barr virus (EBV) were made from affected patients.21 DNA was prepared from all samples; B-cell lines were subjected to RNA extraction and Northern analysis, as previously described.6 Maternal T-cell X-chromosome inactivation patterns were analyzed in somatic cell hybrids, as described,22 or by methylation analysis of the polymorphism in the androgen receptor locus.23 Linkage analysis was performed using tightly linked polymorphic markers flanking IL2RG.24
Polymerase chain reaction (PCR) method.The 8 exons of IL2RG and surrounding genomic sequences were amplified from genomic DNA using primers as shown in Table 1.9 Reactions contained 10 to 100 ng of genomic DNA in 10 to 50 μL with 0.5 μmol/L of each primer, 1.25 U of AmpliTaq polymerase (Perkin Elmer, Norwalk, CT), 200 μmol of each dNTP, 50 mmol/L KCl, 10 mmol/L Tris HCl, pH 8.3, 1.5 mmol/L MgCl2 and 0.001% gelatin. A 94°C incubation for 5 minutes was followed by 35 cycles of 94°C for 45 seconds, an annealing step for 60 seconds, and 72°C for 120 seconds (GeneAmp PCR System 9600; Perkin Elmer). The annealing temperature was progressively decreased from 72°C to 60°C over the first 12 cycles and remained at 60°C thereafter.
SSCP Screening for Mutations in 87 Unrelated Cases of SCID Due to IL2RG Defects
| IL2RG Exon . | Primers Surrounding Exons (forward/reverse) . | Product Size (bp) . | No. of Mutations . | Mutations With Abnormal SSCP . | SSCP Sensitivity (%) . |
|---|---|---|---|---|---|
| 1 | aagctatgacagaggaaacgtg/aggcaccagatctctgtacg | 292 | 6 | 4 | 67 |
| 2 | catttctctttccctccctgc/gagaaaacagtggggtacctgg | 297 | 5 | 4 | 80 |
| 3 | tgcagtacccagattggcc/tccaatgtcccacagtatccc | 291 | 16 | 11 | 69 |
| 4 | ggtattaggggcactaccttcagg/ggccttagctgctacattcacg | 254 | 14 | 13 | 93 |
| 5 | agtagcacagatgacactggtgg/tagaaaggctggggtgttgg | 312 | 23 | 21 | 91 |
| 6 | cagtgcctggcatgtagtagg/accctcctctgctattgtcagc | 225 | 11 | 11 | 100 |
| 7 | tttggtgatggaaggaagcc/acactctgtctgtcttgctggc | 271 | 11 | 7 | 64 |
| 8 | tcctgcccctaattgaccc/cttagggctacaggaccctgg | 276 | 1 | 1 | |
| All exons | 87 | 72 | 83 |
| IL2RG Exon . | Primers Surrounding Exons (forward/reverse) . | Product Size (bp) . | No. of Mutations . | Mutations With Abnormal SSCP . | SSCP Sensitivity (%) . |
|---|---|---|---|---|---|
| 1 | aagctatgacagaggaaacgtg/aggcaccagatctctgtacg | 292 | 6 | 4 | 67 |
| 2 | catttctctttccctccctgc/gagaaaacagtggggtacctgg | 297 | 5 | 4 | 80 |
| 3 | tgcagtacccagattggcc/tccaatgtcccacagtatccc | 291 | 16 | 11 | 69 |
| 4 | ggtattaggggcactaccttcagg/ggccttagctgctacattcacg | 254 | 14 | 13 | 93 |
| 5 | agtagcacagatgacactggtgg/tagaaaggctggggtgttgg | 312 | 23 | 21 | 91 |
| 6 | cagtgcctggcatgtagtagg/accctcctctgctattgtcagc | 225 | 11 | 11 | 100 |
| 7 | tttggtgatggaaggaagcc/acactctgtctgtcttgctggc | 271 | 11 | 7 | 64 |
| 8 | tcctgcccctaattgaccc/cttagggctacaggaccctgg | 276 | 1 | 1 | |
| All exons | 87 | 72 | 83 |
Single-strand conformation polymorphism (SSCP) method.Single exons were amplified with nested primers from the preamplified segments using the PCR conditions described above, with one primer end-labeled with γ-32P-ATP.9 25 PCR products were denatured by boiling, immediately cooled on ice, and separated on nondenaturing Hydrolink MDE gels (FMC, Rockland, ME), with one at 25°C overnight at 7 W and one at 4°C for 4 hours at 45 W.
Sequencing.Templates were IL2RG exons preamplified as described above and isolated from agarose gel bands with a Qiaex kit (Qiagen, Chatsworth, CA) or Microcon 100 spin columns (Amicon, Beverly, MA). A Cyclist DNA sequencing kit (Stratagene, La Jolla, CA) was used with one primer end-labeled with γ-32P-ATP (Redivue; Amersham, Arlington Heights, IL). Mutant sequences were confirmed by forward and reverse sequencing of independently amplified templates from the index patient and, where available, affected or carrier relatives.
Dideoxy fingerprinting.Sequencing reactions were performed as above with dideoxy A (DDA) and dideoxy C (DDC) using IL2RG templates preamplified from patients, carriers or suspected carriers, and controls. Products of these sequencing reactions were denatured by heating to 95°C for 5 minutes, chilled on ice, and separated on nondenaturing MDE gels under the same conditions as for SSCP analysis described above, grouping DDA and DDC lanes.26
Expression and functional studies.B-cell lines from patients were studied for γc expression by cell surface immunofluorescence (IF ) analysis. Cells were incubated with purified rat antihuman γc monoclonal antibody TUGh427 (PharMingen, Los Angeles, CA) either directly labeled with phycoerythrin (PE) or followed by PE-conjugated goat antirat IgG (Caltag Inc, South San Francisco, CA). Cell lines were also stained with anti–IL-2Rα antibody anti-Tac and anti–IL-2Rβ antibodies Mik-β1 and Mik-β2 (all kindly provided by Dr T.A. Waldmann, National Cancer Institute, National Institutes of Health, Bethesda, MD). To assess IL-2 binding in selected B-cell lines, biotin conjugated human IL-2 and avidin-fluorescein were sequentially bound to cells with and without preblocking the biotinylated IL-2 with excess anti–IL-2 antibody (Flourokine Kit; R & D Systems, Minneapolis, MN). Cells were analyzed using a FACSCAN flow cytometer (Becton Dickinson, San Jose, CA).
RESULTS
Frequency of SCID due to X-linked IL2RG mutations.Index affected patients all had a classic, severe infantile SCID phenotype with T lymphocytopenia; those with milder manifestations were excluded. A total of 103 families with a male affected with non–ADA-deficient SCID were evaluated for IL2RG mutations using multiple methods. These methods included pedigree analysis for X-linked inheritance, maternal T-cell X chromosome inactivation pattern, Northern blot analysis, γc cell surface expression in EBV-transformed B-cell lines, and DNA analysis consisting of SSCP, dideoxy fingerprint screening, and direct sequence determination.
IL2RG mutations were eventually ruled out in 16 families by the absence of any abnormality on the above tests and determination of wild-type IL2RG sequence. Sequence analysis performed on DNA from a sample obtained from each of the 16 affected patients before BMT included all 8 exons and flanking regions of IL2RG. These patients were thus presumed to have autosomal non-ADA–deficient SCID. In general, the age at presentation and clinical severity of these 16 patients was not distinguishable from those eventually proven to have X-linked SCID. Two patients categorized as autosomal SCID were recently found to lack Jak3 kinase activity and had elevated percentages of B cells.4 The remaining 14 had SCID of currently unknown etiology. Their immunologic profiles included extremely low numbers of T cells and low mitogen responses. Their B-cell numbers were variable. In 4 cases, B-cell numbers were undetectable and, in the remainder, generally lower than seen in either X-linked SCID or Jak3-deficient SCID patients.4
In the remaining 87 families, X-linked SCID was implicated by one or more of the above indirect criteria and confirmed by finding a significant IL2RG mutation upon sequence analysis. The locations and types of mutations are diagrammed in Fig 1. Available samples from these 87 families included 73 paired samples from an SCID-affected patient and his mother, 12 samples from mothers with deceased affected offspring and/or no available patient sample, and 2 samples from affected males whose mothers declined testing.
IL2RG cDNA map showing exons, cDNA numbers corresponding to the first coding nucleotide of each exon, protein domains, and sites of mutations found in unrelated patients with X-linked SCID in our series. Identical mutations found in unrelated patients are surrounded by shaded boxes. For IL2RG domains: (▨) signal peptide; C, conserved cysteine; W, WSEWS box; TM, transmembrane; B, box1-box2 domain; () 3′ untranslated. For X-linked SCID mutations: (•) point mutation, nonsense; (○) point mutation, missense; (▪) insertion, frame shift; (□) insertion, in frame; (▴) deletion, frame shift; (▵) deletion, in frame; (*) splice site; () site of recurrent mutation.
IL2RG cDNA map showing exons, cDNA numbers corresponding to the first coding nucleotide of each exon, protein domains, and sites of mutations found in unrelated patients with X-linked SCID in our series. Identical mutations found in unrelated patients are surrounded by shaded boxes. For IL2RG domains: (▨) signal peptide; C, conserved cysteine; W, WSEWS box; TM, transmembrane; B, box1-box2 domain; () 3′ untranslated. For X-linked SCID mutations: (•) point mutation, nonsense; (○) point mutation, missense; (▪) insertion, frame shift; (□) insertion, in frame; (▴) deletion, frame shift; (▵) deletion, in frame; (*) splice site; () site of recurrent mutation.
Newly arising IL2RG mutations.An examination of family history showed an X-linked pattern of inheritance in only 33 of the 87 families ultimately shown to have IL2RG mutation (38%); the remaining families presented with a sporadic affected male or two affected brothers, which is consistent with either X-linked or autosomal inheritance. Maternal T-cell X-chromosome inactivation assay indicated whether mothers of SCID boys showed a skewed pattern, as would be expected in a heterozygous carrier of an X-linked IL2RG mutation.28 Direct sequence determination of maternal IL2RG DNA was also performed to verify heterozygosity at the proven IL2RG mutation site. All 25 mothers tested from X-linked pedigrees showed highly skewed T-cell X inactivation, with the SCID-bearing X chromosome being inactive. In addition, all had both mutant and wild-type IL2RG alleles by sequence analysis. However, 12 of the 53 available mothers of sporadic male SCID boys had only wild-type IL2RG sequence in DNA prepared from blood samples, and a normal, random pattern of T-cell X inactivation was correspondingly found in these mothers for whom it was tested. Thus, in our study population, new IL2RG mutations, arising either as maternal mosaicism or within the oocyte donated to the proband, caused 13% (12 of 85 cases with the mother examined) of all X-linked SCID.
Sensitivity of SSCP and DDF in detecting IL2RG mutations.To screen IL2RG exon and surrounding splice sequences rapidly for deviations from wild-type, SSCP analysis was performed. The number of mutations and sensitivity of mutation detection by SSCP in each exon segment is shown in Table 1. Of the 87 mutations ultimately found, 72 (83%) had an SSCP migration pattern at either 4°C or 25°C that was different from that of control DNA. Abnormally migrating bands were detected in no more than one exon for any patient, and no incidental polymorphisms have been seen within the primers designated in greater than 100 X chromosomes from unrelated individuals. As shown in Table 1, our detection rate ranged from around two thirds for exons 1 and 7 to 100% for exon 6.
DDF was also used as a mutation screening tool.26 This method allows detection of conformational changes in strands of increasing lengths from the 5′ to the 3′ end of the segment. Somewhat more laborious than SSCP, DDF was nonetheless successful in detecting migration changes for all 87 of the mutations in our series. Mutations detected by SSCP or by DDF in affected patient DNA were consistently likewise detectable in the heterozygous state in DNA from female carriers.
Recurrent and hot spot mutations in IL2RG.The 87 IL2RG mutations of our unrelated boys with X-linked SCID were not evenly distributed (Table 1 and Fig 1). Exon 5 had 23 or more than one quarter of all the mutations, followed by exon 3 with 16, exon 4 with 14, and exons 6 and 7 with 11 each. We found only a single mutation in exon 8. The patients proved to have 62 different mutations, 52 of which were seen only once in our series and 50 of which have not been published by other groups. Recurrent mutations are marked by shading in Fig 1. Each family sharing a mutation was distinguishable by pedigree and ethnic heritage; moreover, in three instances the new origin of one of the duplicate mutations was proven by sequence and linkage analysis in maternal relatives. The Xq13 chromosomal region of the proband was identified by alleles at tightly linked, highly polymorphic flanking markers, and X chromosomes with the same flanking alleles were traced in his maternal ancestors until one was found with no IL2RG mutation.
Five other mutations were seen 3 or more times, and each of these involved the well-recognized mechanism of cytosine methylation and deamination to thymidine within a cytosine-guanine (CpG) dinucleotide. The region of cDNA 666-691 of IL2RG, just 5′ to the WSEWS motif in exon 5, contains six CpG dinucleotides, the last of which was the site of 11 independent missense mutations, making this the most prominent hot spot for mutation in IL2RG (shaded open circles in exon 5 in Fig 1), as previously noted by our group.10 Five of these mutations were 690C → T, changing arginine 226 to cysteine (R226C). The remaining six, 691G → A, reflected a C to T mutation on the anticoding strand, changing the same arginine to a histidine (R226H). Just six nucleotides 5′ to this hot spot, the mutation 684C → T, causing a nonconservative missense mutation from arginine to tryptophan, R224W, occurred four times in unrelated patients.
Two other hot spot mutation sites were 879C → T, causing a premature termination (solid circles with shading in exon 7 in Fig 1), which was seen six times10; and 868G → A, producing a R285Q missense mutation (shaded open circles) in the last nucleotide of exon 6, which was seen four times. Both cDNA 690 and cDNA 879 mutations have, in addition, been noted in single patients by others.5,12,18 Moreover, a review of published mutations showed a fifth CpG hot spot at cDNA 717 (shaded solid circle in exon 5), producing premature termination, which was seen four times to date by our group and others.16 17
Point mutations in X-linked SCID patients.Thirty-four different point mutations within IL2RG exons were identified, accounting for X-linked SCID in 57 patients (Table 2). Twenty-one patients had 14 different mutations producing premature termination codons, as indicated by solid circles, in exons 2, 3, 4, 5, 7, and 8. One of these, a complex double point mutation in exon 4, changed the L179 codon, TTG, to a termination codon, TAG, while the following codon N180 was also mutated.
Thirty-Four Point Mutations Within Exons in 57 SCID Patients With IL2RG Defects
| IL2RG Exon . | cDNA Mutation . | Protein Mutation . | mRNA Detected* . | γc on Cell Surface by IF† . | Reference‡ . |
|---|---|---|---|---|---|
| 1 | 17G→A [2]ρ | M1I | + | − | |
| 2 | 200T→A | C62•2-154 | − | − | 6 |
| 216G→A | E68K | + | − | 15 | |
| 280A→G | Y89C | + | Trace | 29 | |
| 3 | 355G→A | G114N | + | Trace | 6 |
| 360C→T | Q116• | NA | NA | ||
| 369A→T | K119• | − | − | 5 | |
| 382A→C | H125P | + | Trace | ||
| 387T→A | Y126N | + | + | ||
| 405C→T | Q131• | NA | NA | ||
| 435C→T [2] | Q141• | NA | NA | ||
| 444C→T | Q144• | − | − | 15 | |
| 445A→C | Q144P | + | − | ||
| 4 | 472T→A | I153N | + | + | 6 |
| 499T→A | L162H | NA | NA | 30 | |
| 529T→C | L172P | NA | − | ||
| 529T→A | L172Q | + | − | ||
| 550T→A, 552A→C | L179•, N180H | − | − | ||
| 576C→T | Q188• | − | − | ||
| 581C→A | Y189• | − | − | ||
| 593G→A [2] | W193• | NA | NA | ||
| 5 | 660C→T | Q216• | − | − | |
| 684C→T [4] | R224W | + | − | ||
| 690C→T [5] | R226C | + | Trace | 10, 12, 18 | |
| 691G→A [6] | R226H | + | Trace | 9 | |
| 694T→G | F227C | + | Trace | ||
| 703T→C | L230P | NA | NA | ||
| 708G→A | G232R | NA | NA | ||
| 717C→T | Q235• | − | − | 16, 17 | |
| 736G→T | S241I | NA | NA | 12 | |
| 6 | 823T→G | M270R | + | − | |
| 868G→A [4] | R285Q | + | Trace | 16 | |
| 7 | 879C→T [6] | R289• | + | + | 5, 10 |
| 8 | 978C→T | Q322• | + | + |
| IL2RG Exon . | cDNA Mutation . | Protein Mutation . | mRNA Detected* . | γc on Cell Surface by IF† . | Reference‡ . |
|---|---|---|---|---|---|
| 1 | 17G→A [2]ρ | M1I | + | − | |
| 2 | 200T→A | C62•2-154 | − | − | 6 |
| 216G→A | E68K | + | − | 15 | |
| 280A→G | Y89C | + | Trace | 29 | |
| 3 | 355G→A | G114N | + | Trace | 6 |
| 360C→T | Q116• | NA | NA | ||
| 369A→T | K119• | − | − | 5 | |
| 382A→C | H125P | + | Trace | ||
| 387T→A | Y126N | + | + | ||
| 405C→T | Q131• | NA | NA | ||
| 435C→T [2] | Q141• | NA | NA | ||
| 444C→T | Q144• | − | − | 15 | |
| 445A→C | Q144P | + | − | ||
| 4 | 472T→A | I153N | + | + | 6 |
| 499T→A | L162H | NA | NA | 30 | |
| 529T→C | L172P | NA | − | ||
| 529T→A | L172Q | + | − | ||
| 550T→A, 552A→C | L179•, N180H | − | − | ||
| 576C→T | Q188• | − | − | ||
| 581C→A | Y189• | − | − | ||
| 593G→A [2] | W193• | NA | NA | ||
| 5 | 660C→T | Q216• | − | − | |
| 684C→T [4] | R224W | + | − | ||
| 690C→T [5] | R226C | + | Trace | 10, 12, 18 | |
| 691G→A [6] | R226H | + | Trace | 9 | |
| 694T→G | F227C | + | Trace | ||
| 703T→C | L230P | NA | NA | ||
| 708G→A | G232R | NA | NA | ||
| 717C→T | Q235• | − | − | 16, 17 | |
| 736G→T | S241I | NA | NA | 12 | |
| 6 | 823T→G | M270R | + | − | |
| 868G→A [4] | R285Q | + | Trace | 16 | |
| 7 | 879C→T [6] | R289• | + | + | 5, 10 |
| 8 | 978C→T | Q322• | + | + |
Abbreviation: NA, not available.
By Northern blot.
By cell surface IF staining with TUGh4 monoclonal antibody.
Previous publications, including those by our group, clinical reports and identical mutations found by others.
ρ If multiple cases in our series had the same mutation, the number is shown in brackets.
Solid circle indicates termination codon.
The remaining 20 different point mutations within exons produced single codon changes predicted to cause nonconservative amino acid substitutions, as listed in Table 2.
Insertion and deletion mutations in X-linked SCID patients.Table 3 shows the 14 insertion and deletion mutations we found in IL2RG. Ranging from 1 to 9 nucleotides in size, all were unique, and all but two resulted in frame shifts predicted to give rise to prematurely truncated protein molecules after 1 to 62 missense amino acids. A complex mutation in exon 3 consisted of deletion of one of three adjacent T's at cDNA 398, followed by 6 intact nucleotides and then the deletion of 4 more bases starting at cDNA 406. In this and almost every mutation in this category, the nucleotide sequence immediately adjacent to the deleted or inserted bases contained a recognized motif associated with small deletions.31 Three occurrences of the deletion motif TG(A/G)(A/G)(G/T)(A/C), which were identified by Cooper and Krawczak31 and were noted to be similar to DNA polymerase arrest sites and Ig switch sites, were found in exons 4, 6, and 7. Each was associated with two different mutations in our patients, as listed in Table 3. Moreover, the deletion motif in exon 6 at cDNA 829-834 was the site of two additional deletions published by other groups,12 19 identifying this region as a deletion/insertion hot spot. Other insertions and deletions occurred in a context of palindromic flanking sequences (those at codons L18, W174, N176, Q235, and I273) or short runs of dinucleotide repeats (M145, C231, and V279), polypyrimidines (L42), or polyadenosine (K120).
Fourteen Different Insertions and Deletions in SCID Patients With IL2RG Defects
| IL2RG Exon . | cDNA Mutation . | Nucleotide Sequence Context . | Codon . | Protein Alteration . | mRNA Detected3-150 . | γc on Cell Surface3-151 . | Reference3-152 . |
|---|---|---|---|---|---|---|---|
| 1 | 67delTG | Follows 10-bp inverted repeat | L18 | Frame shift, signal | NA | − | |
| Peptide | |||||||
| 2 | 140delG | Follows 9-bp polypyrimidine | L42 | Frame shift | Trace | − | |
| 3 | 373insA | Within 7-bp poly-A | K120 | Frame shift | − | − | |
| 398delT, 406delAGCT | Complex deletion | F128 | Frame shift | − | − | ||
| 447delA | Follows 2-bp repeat | M145 | Frame shift | − | − | ||
| 4 | 534delTGGAAC | Imperfect double inverted repeat; deletion motif | WN174-5 | Delete 2 amino acids | + | − | |
| 542delCA | Same as above | N176 | Frame shift | − | − | ||
| 5 | 707delTG | Slippage in 2-bp repeat | C231 | Frame shift | − | − | |
| 725duplication9bp | Flanking inverted repeat | QHW235-7 | Repeat 3 amino acids | + | + | 9 | |
| 6 | 833delT | Inverted repeat; del. motif | I273 | Frame shift | NA | NA | |
| 835insAT | Same as above | I274 | Frame shift | + | + | ||
| 851delGT | Slippage in 2-bp repeat | V279 | Frame shift | + | + | ||
| 7 | 922insTA | Del. motif | Y303 | Frame shift | NA | NA | |
| 924delC | Same as above | H304 | Frame shift | + | + |
| IL2RG Exon . | cDNA Mutation . | Nucleotide Sequence Context . | Codon . | Protein Alteration . | mRNA Detected3-150 . | γc on Cell Surface3-151 . | Reference3-152 . |
|---|---|---|---|---|---|---|---|
| 1 | 67delTG | Follows 10-bp inverted repeat | L18 | Frame shift, signal | NA | − | |
| Peptide | |||||||
| 2 | 140delG | Follows 9-bp polypyrimidine | L42 | Frame shift | Trace | − | |
| 3 | 373insA | Within 7-bp poly-A | K120 | Frame shift | − | − | |
| 398delT, 406delAGCT | Complex deletion | F128 | Frame shift | − | − | ||
| 447delA | Follows 2-bp repeat | M145 | Frame shift | − | − | ||
| 4 | 534delTGGAAC | Imperfect double inverted repeat; deletion motif | WN174-5 | Delete 2 amino acids | + | − | |
| 542delCA | Same as above | N176 | Frame shift | − | − | ||
| 5 | 707delTG | Slippage in 2-bp repeat | C231 | Frame shift | − | − | |
| 725duplication9bp | Flanking inverted repeat | QHW235-7 | Repeat 3 amino acids | + | + | 9 | |
| 6 | 833delT | Inverted repeat; del. motif | I273 | Frame shift | NA | NA | |
| 835insAT | Same as above | I274 | Frame shift | + | + | ||
| 851delGT | Slippage in 2-bp repeat | V279 | Frame shift | + | + | ||
| 7 | 922insTA | Del. motif | Y303 | Frame shift | NA | NA | |
| 924delC | Same as above | H304 | Frame shift | + | + |
Abbreviation: NA, not available.
By Northern blot.
By cell surface IF staining with TUGh4 antibody.
Previous publications, including those by our group, clinical reports, and identical mutations found by others.
Splice site mutations in X-linked SCID patients.Fourteen different splice site mutations accounted for 16 unrelated X-linked SCID cases (Table 4). Nine splice mutations were changes in the invariant gt found at the +1 and +2 positions of 5′ splice sites, whereas three changed the +5 g to either c or a. Two 3′ splice site mutations preceded exon 4, at the −1 and −2 invariant ag. The most distinctive splice mutation was a point mutation of the invariant attacking a, or branch point a, at position −15 from exon 3. This mutation occurred independently in two families, one previously reported by our group to have arisen in the germ line of the maternal grandfather of a sporadic case11 and one affecting multiple individuals in a kindred demonstrating more than five generations of X-linked inheritance. Only two other deleterious point mutations of the attacking a nucleotide have been reported in the genetics literature, one causing Spritz β+ thalassemia32 and one associated with the L1 gene in familial X-linked hydrocephalus.33
Fourteen Different Splice Mutations in 16 SCID Patients With IL2RG Defects
| IL2RG Exon . | cDNA Mutation . | Changed Nucleotides (underlined) . | Codon . | mRNA Detected4-150 . | γc Detected4-151 . | References‡ . |
|---|---|---|---|---|---|---|
| 1 | 129(+1) | gtggg→atggg | 39 | NA | NA | |
| 129(+2) [2]ρ | gtggg→gcggg | 39 | Trace, large | − | ||
| 2 | 283(+1) | gtatg→atatg | 90 | NA | NA | |
| 3 | 284(−15) [2] | a→g | 90 | − | − | 11 |
| 468(+1) | gtaat→ataat | 152 | Trace, small | − | 6 | |
| 4 | 469(−1) | ag→aa | 152 | NA | NA | |
| 469(−2) | ag→gg | 152 | NA | NA | ||
| 608(+5) | gtgag→gtgaa | 198 | NA | − | ||
| 6 | 868(+1) | gtgag→ttgag | 285 | Trace | Trace | |
| 868(+2) | del tg; gtgag→g(_)ag | 285 | NA | NA | ||
| 868(+5) | gtgag→gtgac | 285 | NA | NA | ||
| 7 | 938(+1) | gtgag→ttgag | 309 | Trace, large | Trace | |
| 938(+1) | gtgag→atgtg | 309 | Trace, large | Trace | ||
| 938(+5) | gtgag→gtgac | 309 | NA | NA |
| IL2RG Exon . | cDNA Mutation . | Changed Nucleotides (underlined) . | Codon . | mRNA Detected4-150 . | γc Detected4-151 . | References‡ . |
|---|---|---|---|---|---|---|
| 1 | 129(+1) | gtggg→atggg | 39 | NA | NA | |
| 129(+2) [2]ρ | gtggg→gcggg | 39 | Trace, large | − | ||
| 2 | 283(+1) | gtatg→atatg | 90 | NA | NA | |
| 3 | 284(−15) [2] | a→g | 90 | − | − | 11 |
| 468(+1) | gtaat→ataat | 152 | Trace, small | − | 6 | |
| 4 | 469(−1) | ag→aa | 152 | NA | NA | |
| 469(−2) | ag→gg | 152 | NA | NA | ||
| 608(+5) | gtgag→gtgaa | 198 | NA | − | ||
| 6 | 868(+1) | gtgag→ttgag | 285 | Trace | Trace | |
| 868(+2) | del tg; gtgag→g(_)ag | 285 | NA | NA | ||
| 868(+5) | gtgag→gtgac | 285 | NA | NA | ||
| 7 | 938(+1) | gtgag→ttgag | 309 | Trace, large | Trace | |
| 938(+1) | gtgag→atgtg | 309 | Trace, large | Trace | ||
| 938(+5) | gtgag→gtgac | 309 | NA | NA |
Abbreviation: NA, not available.
By Northern blot.
By cell surface IF staining with TUGh4 monoclonal antibody.
Previous publications, including those by our group, clinical reports and identical mutation found by others.
ρ If multiple cases in our series had the same mutation, the number is shown in brackets.
IL2RG mRNA expression.Peripheral blood lymphocytes from all available patients were exposed to EBV-containing supernatants to make transformed B-cell lines. Because the majority of pre-BMT samples contained high percentages of B cells, a typical characteristic of X-linked SCID,4 transformation was usually achieved in samples from these patients. Moreover, in several post-BMT surviving patients, B-cell lines were made that, upon X-linked marker analysis, proved to be of host origin with no detectable donor marker content. B-cell lines for expression studies were thus obtained from 52 patients with X-linked SCID, representing 42 different mutations. Northern blot analysis was used to evaluate IL2RG mRNA expression in these cell lines as compared with greater than 20 control lines that all had a readily detectable 1.8-kb signal (Puck et al6 and data not shown). Where B-cell lines from multiple, unrelated patients with the same mutation were available, mRNA expression among these lines was invariably the same. It was therefore assumed for subsequent population calculations in our series that mRNA expression in patients with no available line would match that of patients with the same mutation for whom a direct measurement was possible. This assumption allowed assignment of mRNA expression for a total of 65 of the 87 X-linked SCID patients.
The results for at least one cell line with each mutation studied are included in Tables 2, 3, and 4. Cell lines with 14 different mutations had undetectable mRNA, and all of these were 5′ to or within exon 5. These mutations included all eight nonsense point mutations with a cell line available through the cDNA 717C → T hot spot (Table 2), an insertion and four frame shift deletions leading to premature terminations at or before cDNA 707 (Table 3), and the intronic attacking a point mutation in intron 2 discussed above.
Trace amounts of IL2RG mRNA were seen in lines bearing six mutations. An unexpected mutation in this category was a frame shift deletion early in exon 2 at cDNA 140. The five other mutations causing trace amounts of mRNA expression were splice mutations leading to premature terminations (Table 4), four of which had mRNA of abnormal size. The splice mutation after exon 3, cDNA 468(+1)g → a, caused skipping of exon 4, whereas the mutations after exons 1, 6, and 7 produced abnormally large IL2RG mRNA with sequence continuing into the introns.
IL2RG mRNA of normal amount and size was found in half (22/42) of the mutations that could be evaluated in B-cell lines, representing 40 unrelated patients. The majority of these mutations were missense point mutations in exons 1 through 6 (Table 2), notably the hot spot mutations at cDNA 690-691 (15 unrelated patients) and cDNA 684 (4 patients). The in-frame deletion and duplication insertion also were associated with normal mRNA (Table 3). Only four mutations predicted to cause premature terminations had normal message levels (frame shift deletions in exon 6, at cDNA 835 and cDNA 851, and in exon 7, at cDNA 924) and two nonsense point mutations in exon 7 at cDNA 879, a hot spot mutated in 6 patients, and in exon 8 at cDNA 978. Because the 22 mutations associated with normal mRNA included frequently mutated hot spot sites, a total of 43 patients (65%) of our X-linked SCID cases whose mutations could be studied by Northern analysis had no detectable abnormality in mRNA expression.
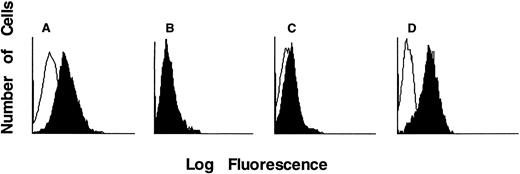
γc protein expression and IL-2 binding.Available B-cell lines were also tested for surface γc protein expression by IF. Strongly positive IF shifts for γc were detected in greater than 50 EBV-transformed B-cell lines from unaffected controls. As noted for mRNA data above, B-cell lines from unrelated patients with the same mutations invariably had similar IF patterns. Three different levels of γc expression, as detected with monoclonal antibody TUGh4, were found (Fig 2). It was expected that no B-cell lines with undetectable message would have γc by IF, and this was indeed the case. For example, as compared with a normal control cell line (Fig 2A), a line bearing the cDNA 576C → T producing a premature stop codon at position 188 in exon 4 showed no shift above background in IF (Fig 2B). Similarly, the two cell lines with cDNA mutation 17G → A, changing the only available starting methionine codon (ATG) to isoleucine (ATA), had no γc expression by IF, even though their mRNA size and amount were normal.
IF staining histograms of EBV B-cell lines incubated with TUGh4 antibody (solid tracings) or no first antibody (open tracings), followed by PE-conjugated antirat IgG. B-cell lines were derived from (A) unaffected control and γc-SCID patients with (B) Q188• terminating in exon 4 with no detectable mRNA, (C) R226H hot spot extracellular point mutation in exon 5, and (D) R289• hot spot intracellular truncation mutation in exon 7.
IF staining histograms of EBV B-cell lines incubated with TUGh4 antibody (solid tracings) or no first antibody (open tracings), followed by PE-conjugated antirat IgG. B-cell lines were derived from (A) unaffected control and γc-SCID patients with (B) Q188• terminating in exon 4 with no detectable mRNA, (C) R226H hot spot extracellular point mutation in exon 5, and (D) R289• hot spot intracellular truncation mutation in exon 7.
Of the 12 cell lines with missense point mutations in the extracellular domain plus the in-frame insertion and deletion mutations, varied IF patterns were seen. Five lines had completely negative IF: E68K in exon 2, Q144P in exon 3, L172Q in exon 4, R224W in exon 5 (Table 2), and the two amino acid deletion of WN174-5 in exon 4 (Table 3). Six lines with missense mutations had a slight, but reproducible, shift in IF: Y89C in exon 2; G114N and H123P in exon 3; and the hot spot mutations R226C and R226H as well as F227C, all in exon 5 (Table 2). An example of the R226H IF pattern is shown in Fig 2C. Only three lines bearing extracellular missense mutations had normal IF, including Y126N in exon 3, I153N in exon 4, and the duplication of three amino acids QHW235-7, including the first W of the WSEWS motif, in exon 5.
Although mutations in and immediately after the transmembrane domain of IL2RG had variable IF staining patterns, those producing intracellular truncation within or after exon 7, regardless of whether they were stop codons, frame shifts, or splice mutations, all had normal IF patterns. An example of IF in a cell line bearing the hot spot R289 mutation to a stop codon is shown in Fig 2D. Thus, intracellular shortening does not affect cell surface expression of γc or its recognition extracellularly by the TUGh4 antibody.
In a subset of lines, IL-2 binding was also tested using fluorescent-labeled IL-2 with or without preblocking by polyclonal goat antihuman IL-2 (not shown). All lines evaluated expressed low but detectable amounts of IL-2Rα and IL-2Rβ chains (not shown). Compared with lines lacking IL2RG mRNA, in which, as expected, minimal IL-2 binding was observed, lines with intracellular truncations showed IL-2 binding comparable to control lines. Interestingly, low IL-2 binding occurred in cell lines with missense point mutations Y89C, H123P, and I153N, whereas IL-2 binding comparable to control lines occurred despite the presence of G114N, Q144P, R226C, and R226H as well as the WN174-5 deletion and the QHW235-7 duplication. Although the IL-2 binding assay is not quantitative and could be subject to variable expression of IL-2 receptor chains in EBV-transformed cells, the discordance between TUGh4 staining and IL-2 binding is of interest. Although the binding epitope on γc for TUGh4 has not been reported, this antibody is known not to block cytokine binding by γc.26 The studies described above suggest that Q144 or WN174-5 could be important for display of the TUGh4 epitope, whereas the mutations at these sites do not interfere with IL-2 binding.
DISCUSSION
We have found 87 specific X-linked SCID-causing mutations in and adjacent to all eight IL2RG exons from male patients referred from throughout North America. Although no single mutation predominated, the types and locations of mutations were not evenly distributed. No large DNA deletions or rearrangements occurred in our series, although two instances of large deletions have been reported from England16 and France.20 In addition, no evidence for regulatory mutations was found in the patients in this series in that all who had decreased or absent IL2RG mRNA had mutations identified within the exons and splice regions. Two thirds of our patients had missense or nonsense point mutations, with the remainder being equally divided between splice mutations and insertions and deletions of 1 to 9 nucleotides. Although nearly two thirds of our mutations were unique (54/87), the others were seen repeatedly in unrelated kindreds. We have now defined five CpG dinucleotide hot spots, which together account for 30% of the mutations we studied. Several unique mutations were clustered near repeated nucleotides and deletion motifs in exons 3, 4, and 5, and an insertion/deletion hot spot was identified in exon 6.
The relative frequency of X-linked SCID in comparison with the autosomal SCID syndromes of known and unknown genotype has not been determined in North America. A striking male predominance in all studies indicates that X-linked SCID is the most frequent form. In our series of 103 families with males with non-ADA–deficient SCID, only 16 proved not to have a deleterious mutation in the IL2RG gene encoding γc. If one assumes that 16 females might also have been diagnosed with autosomal SCID during the period of this study, a rough estimate for X-linked SCID frequency of 73% of all non-ADA–deficient SCID is reached. This number is higher than past estimates from series from the United States2 and Europe34 before the availability of exhaustive detection methods for IL2RG mutation. Our study population could have a bias of ascertainment; however, this estimate is not greatly different from the maximum frequency estimate for X-linked SCID of 66% of non-ADA–deficient SCID found by Buckley et al4 in 108 consecutive SCID patients, partially overlapping with those in this study, evaluated at the Duke University Medical Center.
The 16 SCID male patients in the current study without IL2RG mutation had a classical, severe phenotype and, for those with available data, low numbers of T cells and variable numbers of B cells (undetectable to elevated) and NK cells (undetectable to normal). Recently, 2 of these patients were found to be deficient in Jak3 kinase activity.4,35,36 The other SCID patients in this group have no specific genotype assignments to date, although those with no B cells are candidates for deficiency of the V(D)J recombinase enzymes RAG-1 or RAG-2, as recently reported by Schwartz et al.37
Although it has previously been appreciated that newly arising IL2RG mutations cause a significant proportion of X-linked SCID,38 this fact was underlined in the present series. Not only did 62% of mutation-proven IL2RG-deficient SCID cases have no family history documenting X-linked mutation transmission, but 13% of all the cases with mother and son available for study (a full 23% of sporadic SCID cases) had mothers who failed to show skewed X chromosome inactivation in peripheral blood lymphocytes. This relatively high proportion of IL2RG mutations arising in the maternal germline complicates genetic counseling in this disease. First, random maternal X chromosome inactivation or other tests for IL2RG mutation in maternal blood cannot rule out X-linked SCID in families in which a sporadic male proband is deceased. Moreover, the recurrence risk for a mother of a sporadic X-linked SCID male cannot be determined in this setting. If the affected male's X chromosome became mutated only in the oocyte donated to him, his mother's risk of future affected offspring would be zero. However, mutation in a germ cell progenitor, as has already been documented for X-linked SCID,9 could mean that the mother's recurrence risk would be as high as 50% for each male pregnancy.
Detection of mutations causing X-linked SCID also remains a challenge. Northern blots would show abnormalities in only 35% of patients, based on the IL2RG mutations in cases we were able to test. Cell surface staining of patient-derived B-cell lines with an anti-γc antibody was somewhat more sensitive, showing no γc staining in 47% and trace amounts in 32%, but normal IF intensity in 21% of patients, again based on mutations with cell lines available. DNA-based methods offer the advantages of mutation detection from biopsy or autopsy material, rather than requiring patient pretransplant blood samples. Maternal relatives can serve as the source of DNA bearing the IL2RG mutation to be sought, except in cases of new mutation in their offspring. Moreover, specific mutations at the sequence level are the most accurate means of prenatal and carrier diagnosis within families.
SSCP has proven a valuable screening technique for localizing IL2RG mutations, but its overall sensitivity of only 83% in this series, which is very close to the 85% reported by Clark et al,16 precluded its use alone for identifying mutations. The variation in SSCP sensitivity from 64% in exon 7 to 100% in exon 6 reflected primary sequence content as well as the predicted inverse relationship between sensitivity and fragment size.39 Indeed, even in the largest segment, 312 bp including exon 5, SSCP sensitivity was greater than 91%, detecting 21 of 23 mutations. Exon 5 was also the exon with the highest number of mutations. The specific mutations missed by SSCP screening were all single base substitutions, and 11 of the 15 were in splice sequences or the initiating methionine codon, all occurring near the ends of the segments amplified by the primer sets used. DDF, although somewhat more time consuming than SSCP, successfully detected all of the IL2RG mutations we have encountered.
A sufficient number of IL2RG mutations have been accumulated so that correlations between mutation type and functional consequences can be made. In X-linked SCID cases with mutations leading to premature termination in exons 1 through 5, mRNA levels were, with one exception, undetectable by Northern blot. IL2RG is thus typical of many well-studied genes in which mutations producing early termination lead to unstable mRNA. However, point mutations and in-frame insertions and deletions throughout all exons (open symbols in Fig 1) may express abnormal γc chains, and the location and type of these mutations may lead to better understanding of functional domains of the protein. Mutations in exons 2 and 3 involving or adjacent to the four cysteine residues conserved in all cytokine receptor family proteins are expected to disrupt γc configuration, whereas similar detrimental effects are predictable for mutations in exon 5 in or near the canonical juxtamembrane WSEWS motif. Interestingly, the open-symbol mutations in distal exons 3 and 4 (Fig 1) do not affect known functional motifs. Among these mutations are the amino acid substitution at Q144P and the deletion of two amino acids WN174-5, which resulted in loss of binding by the anti-γc antibody TUGh4, while nonetheless preserving IL-2 binding capability. Further study of these extracellular mutations is expected to shed light on the currently poorly understood interactions of γc with its various cytokine receptor partners as well as with cytokine molecules themselves.
The transmembrane domain of IL2RG occupies most of exon 6. Although mRNA with some mutations in this region may be stable enough for protein transcription to occur, disruption of either the hydrophobic transmembrane amino acids or the anchoring residues in the immediate cytoplasmic region could result in γc protein chains unlikely to be stably expressed on the cell surface.
In general, mutations producing premature truncations in exons 7 and 8 have been found to express mRNA levels in EBV-transformed B-cell lines comparable to the levels seen in EBV lines from normal controls. The proximal intracellular portion of γc contains an SH2 or Box-1/Box-2 homologous domain between amino acids 288 and 300 (B in Fig 1), which is common to cytokine receptors and other transmembrane signaling molecules. This domain is important for interaction with the intracellular signaling kinase Jak3.7,40 However, the considerably more distal truncation mutation at cDNA 978 in exon 8 in one of our patients with typical SCID confirms in vitro data7 8 showing that the terminal 40 amino acids of γc are critical for proper association with Jak3.
Correlations in this series of X-linked SCID patients between genotypes and clinical phenotypes have not yet been informative, in part because the study was designed to enroll patients with severe, infantile presentation. We did not find either of the two IL2RG mutations reported to cause a milder combined immunodeficiency presenting later in life.7,13 Interestingly, of 5 patients sharing the same R226C hot spot mutation, 1 had an atypically low B-cell number for X-linked SCID of 75/μL, whereas another had an atypical normal number of NK cells.10 Thus environmental factors or other genetic loci may influence the immunologic phenotype. Nonetheless, recognition of the diversity of mutations may become important in the selection of patients for gene therapy trials using retroviral insertion of normal IL2RG into autologous hematopoeitic progenitor cells. We have found that as many as two thirds of X-linked SCID patients may express defective γc chains at levels comparable to the expression of wild-type chains in normal individuals. Some, or perhaps many, of the defects in these endogenous proteins may cause them to interfere with normal γc even if it can be introduced and expressed in the patients' lymphoid progenitors. Further studies will be needed to clarify which patients may be the most appropriate gene therapy candidates.
ACKNOWLEDGMENT
The authors thank the immunologists, geneticists, and genetic counselors and families whose participation has made these studies possible, including L. Middelton and J. Davis and Drs R. Hostoffer, J. Innes, S. Josephs, C. Cunningham-Rundles, K. Sullivan, and R. Schiff. C. Goldman helped us with IF analysis and expert assistance was provided by S. Schiff.
Supported by March of Dimes Grant No. FY92-0892 to J.M.P.; by National Institutes of Health Grants No. HD23679 to J.M.P., 5R37-AI18613 and 1U19-AI38550 to R.H.B., and 9-PO1-AI32918 to T.N.S.; by The David Fund; by Texas Children's Hospital; and by American Heart Association Grant No. 88G-194 to H.M.R.
Address reprint requests to Jennifer M. Puck, MD, NCHGR/NIH, Bldg 49, Room 3W14, 49 Convent Dr, Bethesda, MD 20892-4442.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal