Abstract
Loss of chromosome 7 (−7) or deletion of its long arm (7q−) are recurring chromosome abnormalities in myeloid disorders, especially in therapy-related myelodysplastic syndrome (t-MDS) and acute myeloid leukemia (t-AML). The association of −7/7q− with myeloid leukemia suggests that these regions contain a novel tumor suppressor gene(s) whose loss of function contributes to leukemic transformation or tumor progression. Based on chromosome banding analysis, two critical regions have been identified: one in band 7q22 and a second in bands 7q32-q35. We analyzed bone marrow and blood samples from 21 patients with myeloid leukemia (chronic myeloid leukemia, n = 2; de novo MDS, n = 4; de novo AML, n = 13; t-AML, n = 2) that on chromosome banding analysis exhibited deletions (n = 19) or reciprocal translocations (n = 2) of band 7q22 using fluorescence in situ hybridization. As probes, we used Alu-polymerase chain reaction products from 22 yeast artificial chromosome (YAC) clones that span chromosome bands 7q21.1-q32, including representative clones from a panel of YACs recognizing a contiguous genomic DNA fragment of 5 to 6 Mb in band 7q22. In the 19 cases with deletions, we identified two distinct commonly deleted regions: one region within band 7q22 was defined by the two CML cases; the second region encompassed a distal part of band 7q22 and the entire band 7q31 and was defined by the MDS/AML cases. The breakpoint of one of the reciprocal translocations was mapped to 7q21.3, which is centromeric to both of the commonly deleted regions. The breakpoint of the second translocation, which was present in unstimulated bone marrow and phytohemagglutinin-stimulated blood of an MDS patient, was localized to a 400-kb genomic segment in 7q22 within the deletion cluster of the MDS/AML cases. In conclusion, our data show marked heterogeneity of 7q22 deletion and translocation breakpoints in myeloid leukemias, suggesting the existence of more than one pathogenetically relevant gene.
LOSS OF CHROMOSOME 7 (−7) or deletion of its long arm (7q−) are recurring chromosome abnormalities in myeloid disorders. These aberrations are associated with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), in particular with therapy-related MDS/AML (t-MDS/t-AML) after therapy with alkylating agents or secondary MDS/AML after occupational exposure to chemical mutagens.1-3 Furthermore, −7/7q− occur in MDS and AML that develop in patients with constitutional disorders (eg, Fanconi's anemia, Kostmann's syndrome, neurofibromatosis type 1, and familial monosomy 7).4 Clinically, myeloid leukemias exhibiting −7/7q− are associated with high susceptibility to infections, poor response to chemotherapy, and short survival times.1 5
Based on chromosome banding analysis, there appear to be two critical regions on the long arm of chromosome 7: one in band 7q22 and a second in bands 7q32-q35.3,6-9 The recurrent loss of chromosome material suggests that these regions contain a novel tumor suppressor gene(s) whose loss of function leads to leukemic transformation. At present, there are scarce data on the molecular characterization of the genomic region affected by these deletions. No tumor suppressor gene of pathogenic significance has been identified. Kere et al10,11 studied 4 patients with myeloid disorders exhibiting partial 7q deletions. Using restriction fragment length polymorphism (RFLP) and quantitative Southern blot analysis, they localized the proximal breakpoints of all 4 cases between the genes for erythropoietin (EPO) and plasminogen activator inhibitor 1 (PLANH1).10 All deletions were interstitial, with variability of the distal breakpoints.11 In another RFLP study of 5 patients with myeloid disorders and 7q deletions, all 5 deletions were interstitial, with great variability of the proximal and distal breakpoints.12 However, in these studies, the resolution of the deletion map was limited because the applied probes were scattered along a large genomic region.
Recently, fluorescence in situ hybridization (FISH) with mapped cosmid and yeast artificial chromosome (YAC) clones has been used for the molecular analysis of chromosome 7 aberrations.9,13 Johnson et al13 defined a small genomic region in a patient carrying a constitutional inversion inv(7)(q22.1q34) and in an unrelated patient with a translocation t(2; 7)(p11; q21.2) who both had developed MDS. The breakpoints of both the inversion and the translocation were localized to a 650-kb genomic fragment in band 7q22.1. In a series of 15 patients with myeloid or lymphoid malignancies, Le Beau et al9 recently described a 2- to 3-Mb commonly deleted region in band 7q22 that is located distally to the inversion breakpoint described by Johnson et al.13
In the present study, we characterized deletions and translocations affecting chromosome band 7q22 in 21 patients with myeloid leukemias using FISH. As probes, we selected 22 clones from a contig map of YAC clones encompassing bands 7q21.1-q32.14-17 Because overlapping YAC clones were used, it was possible to systematically delineate the extent of the deletions and to locate the breakpoints of reciprocal translocations on the molecular level.
MATERIALS AND METHODS
Patients
Bone marrow and/or blood samples from 21 adult patients with myeloid disorders that on banding analysis exhibited deletions or translocations of chromosome band 7q22 were analyzed. The diagnosis were as follows: chronic-phase chronic myeloid leukemia (CML), n = 2 (nos. 1 and 2); de novo MDS, n = 4 (nos. 3 through 6); de novo AML, n = 13 (nos. 7 through 19); and t-AML, n = 2 (nos. 20 and 21). G-banding analysis was performed using standard methods. Images were captured and processed in the Ikaros system (MetaSystems, Altlussheim, Germany) and the karyotypes were designated according to the International System for Cytogenetic Nomenclature.18
DNA Probes
For the interphase and metaphase FISH experiments, we selected the following 22 YAC clones (Table 1) that were previously mapped to chromosome bands 7q21.1 to 7q32 (genes that are present on each YAC are given in parentheses): HSC7E520 (CACNL2A; HGF ), HSC7E590, HSC7E481 (PGY1, PGY3), HSC7E502, HSC7E465 (CALCR), HSC7E783 (ASNS), HSC7E911, HSC7E808, HSC7E914, HSC7E737 (PLANH1), HSC7E791 (CUTL1), HSC7E329, HSC7E316, HSC7E441, HSC7E485, HSC7E506, HSC7E572, HSC7E161 (PRKAR2B, DRA), HSC7E132, HSC7E4 (CAPZA2, MET), HSC7E222, and HSC7E1289 (GRM8).14-17,19 20 Information on the YAC clones is available at http://www.genet.sickkids.on.ca/chromosome7/
Designation of YAC Clones, Chromosomal Localization, Size, and DNA Markers/Genes on Each YAC Clone
| Name . | Location . | Size (kb) . | DNA Markers . |
|---|---|---|---|
| HSC7E520 | 7q21.1 | 500 | CACNL2A, HGF, D7S581, D7S802, D7S849, D7S2275E |
| HSC7E590 | 7q21.1-q21.2 | 460 | D7S166, D7S195, D7S260, D7S524, D7S644, D7S2537 |
| HSC7E481 | 7q21.1-q21.3 | 900 | PGY1, PGY3, D7S269, D7S575, D7S1642, D7S1646, D7S2621, D7S2623, D7S2624, D7S2626, D7S2628, D7S2786, Cdy11e05 |
| HSC7E502 | 7q21.1-q21.3 | 600 | D7S183, D7S492, D7S569, D7S570, D7S577 |
| HSC7E465 | 7q21.3 | 700 | CALCR, D7S15, D7S652, D7S657, D7S2430, HSC7E1275RV |
| HSC7E783 | 7q21.3-q22.1 | 650 | ASNS, D7S82, D7S554, HSC7E1131RV, UT901 |
| HSC7E911 | 7q22 | 580 | D7S651, NIB217, 932D8.1, E911.1 |
| HSC7E808 | 7q22 | 470 | D7S647, D7S2498, D7S2832, AFMa163ya9, E808.3, CYP3A4 |
| HSC7E914 | 7q22 | 340 | D7S647, D7S2832, AFMa163ya9, CYP3A4 |
| HSC7E737 | 7q22 | 250 | PLANHI, D7S477, UT682 |
| HSC7E791 | 7q22 | 250 | CUTL1, D7S515, D7S666, D7S740, D7S2433, D7S2536 |
| HSC7E329 | 7q22 | 380 | D7S2448, APCR.HSC7E452, APCR.HSC7E570, 942E8.18 |
| HSC7E316 | 7q22 | 440 | APCR.HSC7E452, 942E8.18 |
| HSC7E441 | 7q22 | 480 | D7S76, D7S238, D7S561, D7S658, D7S796, D7S818, D7S2504 |
| HSC7E485 | 7q22 | 540 | D7S76, D7S238, D7S561, D7S562, D7S563, D7S796, D7S818 |
| HSC7E506 | 7q22 | 520 | D7S240, D7S561, D7S562, D7S563, D7S572, D7S579, D7S584, D7S2509 |
| HSC7E572 | 7q22.3-q31.1 | 400 | D7S1493, D7S1799, D7S2494 |
| HSC7E161 | 7q22.3-q31.1 | 450 | PRKAR2B, DRA, D7S496, D7S2317, D7S2459, Bdy96E06 |
| HSC7E132 | 7q22.3-q31.1 | 650 | D7S13, D7S1763, Bdy31e11 |
| HSC7E4 | 7q31.2-q31.3 | 600 | CAPZA2, MET, D7S567, D7S568, D7S1764, D7S2460 |
| HSC7E222 | 7q31.3-q32.1 | 580 | D7S480, D7S650, D7S685, D7S2821, Cdy0rd04 |
| HSC7E1289 | 7q32.1 | 1800 | GRM8, D7S514, D7S680, D7S686, D7S1498, D7S1801, D7S1822, D7S1873, D7S1874, D7S2244 |
| HSC7E526 | 7q36 | 560 | D7S68, D7S593, D7S594 |
| Name . | Location . | Size (kb) . | DNA Markers . |
|---|---|---|---|
| HSC7E520 | 7q21.1 | 500 | CACNL2A, HGF, D7S581, D7S802, D7S849, D7S2275E |
| HSC7E590 | 7q21.1-q21.2 | 460 | D7S166, D7S195, D7S260, D7S524, D7S644, D7S2537 |
| HSC7E481 | 7q21.1-q21.3 | 900 | PGY1, PGY3, D7S269, D7S575, D7S1642, D7S1646, D7S2621, D7S2623, D7S2624, D7S2626, D7S2628, D7S2786, Cdy11e05 |
| HSC7E502 | 7q21.1-q21.3 | 600 | D7S183, D7S492, D7S569, D7S570, D7S577 |
| HSC7E465 | 7q21.3 | 700 | CALCR, D7S15, D7S652, D7S657, D7S2430, HSC7E1275RV |
| HSC7E783 | 7q21.3-q22.1 | 650 | ASNS, D7S82, D7S554, HSC7E1131RV, UT901 |
| HSC7E911 | 7q22 | 580 | D7S651, NIB217, 932D8.1, E911.1 |
| HSC7E808 | 7q22 | 470 | D7S647, D7S2498, D7S2832, AFMa163ya9, E808.3, CYP3A4 |
| HSC7E914 | 7q22 | 340 | D7S647, D7S2832, AFMa163ya9, CYP3A4 |
| HSC7E737 | 7q22 | 250 | PLANHI, D7S477, UT682 |
| HSC7E791 | 7q22 | 250 | CUTL1, D7S515, D7S666, D7S740, D7S2433, D7S2536 |
| HSC7E329 | 7q22 | 380 | D7S2448, APCR.HSC7E452, APCR.HSC7E570, 942E8.18 |
| HSC7E316 | 7q22 | 440 | APCR.HSC7E452, 942E8.18 |
| HSC7E441 | 7q22 | 480 | D7S76, D7S238, D7S561, D7S658, D7S796, D7S818, D7S2504 |
| HSC7E485 | 7q22 | 540 | D7S76, D7S238, D7S561, D7S562, D7S563, D7S796, D7S818 |
| HSC7E506 | 7q22 | 520 | D7S240, D7S561, D7S562, D7S563, D7S572, D7S579, D7S584, D7S2509 |
| HSC7E572 | 7q22.3-q31.1 | 400 | D7S1493, D7S1799, D7S2494 |
| HSC7E161 | 7q22.3-q31.1 | 450 | PRKAR2B, DRA, D7S496, D7S2317, D7S2459, Bdy96E06 |
| HSC7E132 | 7q22.3-q31.1 | 650 | D7S13, D7S1763, Bdy31e11 |
| HSC7E4 | 7q31.2-q31.3 | 600 | CAPZA2, MET, D7S567, D7S568, D7S1764, D7S2460 |
| HSC7E222 | 7q31.3-q32.1 | 580 | D7S480, D7S650, D7S685, D7S2821, Cdy0rd04 |
| HSC7E1289 | 7q32.1 | 1800 | GRM8, D7S514, D7S680, D7S686, D7S1498, D7S1801, D7S1822, D7S1873, D7S1874, D7S2244 |
| HSC7E526 | 7q36 | 560 | D7S68, D7S593, D7S594 |
The region between YAC clones HSC7E808 to HSC7E572 represents approximately 5 to 6 Mb of DNA and this region is covered completely by an overlapping set of YAC clones.17 HSC7E914, HSC7E737, HSC7E791, HSC7E329, HSC7E316, HSC7E441, HSC7E485, and HSC7E506 are representative clones from this contig that were chosen to be uniformly spaced throughout the region. The genes encoding EPO and ACHE are located between clones HSC7E911 and HSC7E808.20 To mark the end of the chromosome, we used clone HSC7E526, which begins approximately 100 kb from the end of the chromosome and extends 560 kb towards the centromere.20
Human sequences from the YAC clones were generated by a polymerase chain reaction (PCR) protocol using primers directed against Alu-sequences.21 Amplification was performed in a 100 μL reaction mixture containing approximately 160 ng YAC-DNA, 100 mmol/L of the four dNTPs (Boehringer Mannheim, Mannheim, Germany), 10 μL PCR buffer (Boehringer Mannheim), and 2.0 mmol/L MgCl2 (Boehringer Mannheim). Three Alu-PCR reactions were performed using either the primers CL1, CL2, or a combination of both (0.5 μmol/L). The products of all three reactions were combined for the FISH experiments. The probes were labeled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP (Boehringer Mannheim).
FISH
FISH was performed as described.22,23 The hybridization mixture contained approximately 250 ng labeled YAC-DNA, 10 μg Cot-1 DNA fraction (BRL/Life Technologies, Gaithersburg, MD), and 10 μg herring sperm DNA (Boehringer Mannheim).
Interphase cytogenetic analysis was performed for deletion mapping. To monitor the hybridization efficiency, we cohybridized with a YAC clone from 7q that mapped outside the region of interest. By analogy to our previous studies on deletion analyses in B-cell chronic lymphocytic leukemia, the cut-off level was defined by the mean + 3 standard deviations (SD) of the frequency of control cells exhibiting only one fluorescence signal.24 25 The cut-off levels were determined for 5 representative YACs from the YAC map (range, 4.5% to 7.6%). Signal numbers were enumerated in 200 to 300 nuclei. The breakpoints of the two balanced translocations were identified by metaphase FISH. Images were captured using a cooled CCD camera (Xillix Technologies Corp, Richmond, British Columbia, Canada).
RESULTS
G-Banding
The complete karyotypes of the 21 leukemias are given in Table 2.
Karyotypes of the 21 Myeloid Leukemias
| Tumor . | Karyotype . |
|---|---|
| CML | |
| 1 | 46,XX,t(9; 22)(q34;q11)[8]/46,XX,del(7)(q11.2q22),t(9; 22)(q34;q11)[7] |
| 2 | 46,XY,t(9; 22)(q34;q11)[7]/46,XX,del(7)(q22q22),t(9; 22)(q34;q11)[3] |
| MDS | |
| 3 | 46,XY[7]/46,XY,del(7)(q22)[3] |
| 4 | 46,XX[3]/46,XX,add(6)(p21),del(7)(q22q36)[7] |
| 5 | 46,XX,del(7)(q22)[10] |
| 6* | 46,XY,t(3; 7)(p13;q22)[20] |
| De novo AML | |
| 7 | 47,XY,del(7)(q22q36),+8[20] |
| 8 | 46,XY,del(7)(q11q36),?t(8; 19)(q24;q11),del(9)(q11q22), add(21)(p11)[5]/46,XY[15] |
| 9 | 48,XY,del(3)(p21),+4,der(5)t(5; 13)(q31;q12 or 14),del(7)(q22),−13,+22,+mar[20] |
| 10 | 46,XY,del(7)(q22q32)[15] |
| 11 | 50,XY,der(5)t(1; 5)(p22;q13),del(7)(q22),+11,−13, add(16)(q2?),−21,+5mar[12] |
| 12 | 47,XY,−5,del(7)(q21),+8,+r(11q?),−13,add(17)(p11), +mar[15] |
| 13 | 45,XY,add(2)(q3?),del(3)(q21),−5,del(7)(q2?1),der(13; 21)(q10;q10),−17,add(21)(p11),+r,inc[6] |
| 14 | 46,XX,t(2; 11)(q37;q23),del(7)(q11)[21] |
| 15 | 45,Y,del(X)(p11),−2,add(3)(q2?7),add(3)(p21),del(4) (q21q31),−5,del(7)(q22q32),add(8)(q2?4),del(9) (p22),−13,add(16)(p13),der(17)t(2; 17)(q21;p11), +mar1,+mar2[16] |
| 16 | 49,XY,del(7)(q22),+8,t(8; 16)(p11;p13),+13,+13[15] |
| 17† | No evaluable metaphases |
| 18 | 46,XX,+del(1)(p22),del(7)(q11q32 or 34),−13[15] |
| 19 | 46,XX[7]/46,XX,t(3; 7)(q28;q22)[22] |
| t-AML | |
| 20 | 40,XY,−5,−6,add(7)(q?),−9,add(12)(p11or13),+13, −14,−15,−16,−16,der(?)t(?; 17)(?;q21),−20,−22, +mar1,+mar2,inc[5] |
| 21 | 46,XX,del(3)(q21q27),add(7)(q22),add(19)(p13)[15] |
| Tumor . | Karyotype . |
|---|---|
| CML | |
| 1 | 46,XX,t(9; 22)(q34;q11)[8]/46,XX,del(7)(q11.2q22),t(9; 22)(q34;q11)[7] |
| 2 | 46,XY,t(9; 22)(q34;q11)[7]/46,XX,del(7)(q22q22),t(9; 22)(q34;q11)[3] |
| MDS | |
| 3 | 46,XY[7]/46,XY,del(7)(q22)[3] |
| 4 | 46,XX[3]/46,XX,add(6)(p21),del(7)(q22q36)[7] |
| 5 | 46,XX,del(7)(q22)[10] |
| 6* | 46,XY,t(3; 7)(p13;q22)[20] |
| De novo AML | |
| 7 | 47,XY,del(7)(q22q36),+8[20] |
| 8 | 46,XY,del(7)(q11q36),?t(8; 19)(q24;q11),del(9)(q11q22), add(21)(p11)[5]/46,XY[15] |
| 9 | 48,XY,del(3)(p21),+4,der(5)t(5; 13)(q31;q12 or 14),del(7)(q22),−13,+22,+mar[20] |
| 10 | 46,XY,del(7)(q22q32)[15] |
| 11 | 50,XY,der(5)t(1; 5)(p22;q13),del(7)(q22),+11,−13, add(16)(q2?),−21,+5mar[12] |
| 12 | 47,XY,−5,del(7)(q21),+8,+r(11q?),−13,add(17)(p11), +mar[15] |
| 13 | 45,XY,add(2)(q3?),del(3)(q21),−5,del(7)(q2?1),der(13; 21)(q10;q10),−17,add(21)(p11),+r,inc[6] |
| 14 | 46,XX,t(2; 11)(q37;q23),del(7)(q11)[21] |
| 15 | 45,Y,del(X)(p11),−2,add(3)(q2?7),add(3)(p21),del(4) (q21q31),−5,del(7)(q22q32),add(8)(q2?4),del(9) (p22),−13,add(16)(p13),der(17)t(2; 17)(q21;p11), +mar1,+mar2[16] |
| 16 | 49,XY,del(7)(q22),+8,t(8; 16)(p11;p13),+13,+13[15] |
| 17† | No evaluable metaphases |
| 18 | 46,XX,+del(1)(p22),del(7)(q11q32 or 34),−13[15] |
| 19 | 46,XX[7]/46,XX,t(3; 7)(q28;q22)[22] |
| t-AML | |
| 20 | 40,XY,−5,−6,add(7)(q?),−9,add(12)(p11or13),+13, −14,−15,−16,−16,der(?)t(?; 17)(?;q21),−20,−22, +mar1,+mar2,inc[5] |
| 21 | 46,XX,del(3)(q21q27),add(7)(q22),add(19)(p13)[15] |
Translocation found in unstimulated bone marrow and PHA-stimulated blood.
7q− identified by interphase cytogenetics using clone HSC7E506 (see Fischer et al23 ).
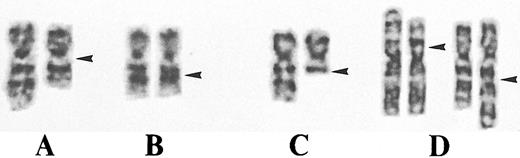
CML.Both CML cases were studied at diagnosis and exhibited interstitial deletions of band 7q22 in a subclone. Case no. 1 had a proximal deletion del(7)(q11.2q22) (Fig 1A). Case no. 2 had a small deletion del(7)(q22q22) within band 7q22 (Fig 1B).
Representative partial G-banding karyotypes. (A) CML (case no. 1) exhibiting an interstitial deletion del(7)(q11.2q22). (B) CML (case no. 2) with a small interstitial deletion del(7)(q22q22). Note that bands 7q21 and 7q31 are closer to each other in the deleted than in the normal chromosome 7 homolog. (C) MDS (case no. 3) with a terminal deletion del(7)(q22). (D) MDS (case no. 6) exhibiting a reciprocal translocation t(3; 7)(p13; q22) in unstimulated bone marrow and PHA-stimulated blood.
Representative partial G-banding karyotypes. (A) CML (case no. 1) exhibiting an interstitial deletion del(7)(q11.2q22). (B) CML (case no. 2) with a small interstitial deletion del(7)(q22q22). Note that bands 7q21 and 7q31 are closer to each other in the deleted than in the normal chromosome 7 homolog. (C) MDS (case no. 3) with a terminal deletion del(7)(q22). (D) MDS (case no. 6) exhibiting a reciprocal translocation t(3; 7)(p13; q22) in unstimulated bone marrow and PHA-stimulated blood.
MDS/AML.Of the 4 cases with MDS, 3 (nos. 3 through 5) had large 7q deletions; all proximal breakpoints involved band 7q22 (Fig 1C). The remaining case (no. 6) had a balanced translocation t(3; 7)(p13; q22) that was present in unstimulated bone marrow and phytohemagglutinin (PHA)-stimulated blood (Fig 1D). Of the 15 cases with de novo or t-AML, 14 (nos. 7 through 18, 20, and 21) had terminal or interstitial deletions that involved or encompassed band 7q22. The smallest deletions were identified in cases no. 10 [del(7)(q22q32)] and 15 [del(7)(q22q32)], the latter within a highly complex karyotype. One AML case (no. 17) had no evaluable metaphase spreads on chromosome banding analysis; in this case, the deletion was identified by interphase cytogenetics with YAC clone HSC7E506.23 The remaining case (no. 19) had a balanced translocation t(3; 7)(q28; q22).
Deletion and Translocation Mapping by FISH
The results of the deletion/translocation mapping by FISH are given in Fig 2. In the 19 cases of CML, MDS, and AML containing deletions, we identified two distinct commonly deleted regions.
Mapping of deletions and translocations involving chromosome band 7q22 in 21 myeloid leukemias by FISH. del, deletion (only 1 fluorescence signal); 3, three fluorescence signals indicating the translocation breakpoint; di, disomy (2 fluorescence signals indicating retention of both alleles); empty boxes, not done. Light grey boxes indicate the extent of the deletion. Dark grey boxes indicate the two commonly deleted segments in CML and MDS/AML, respectively. *Clones HSC7E808 through HSC7E572 recognize a contiguous genomic fragment in chromosome band 7q22.
Mapping of deletions and translocations involving chromosome band 7q22 in 21 myeloid leukemias by FISH. del, deletion (only 1 fluorescence signal); 3, three fluorescence signals indicating the translocation breakpoint; di, disomy (2 fluorescence signals indicating retention of both alleles); empty boxes, not done. Light grey boxes indicate the extent of the deletion. Dark grey boxes indicate the two commonly deleted segments in CML and MDS/AML, respectively. *Clones HSC7E808 through HSC7E572 recognize a contiguous genomic fragment in chromosome band 7q22.
CML.The commonly deleted segment in the 2 CML cases extended from YAC clones HSC7E914 to HSC7E791 (Fig 2). This segment was defined by case no. 2, which on banding analysis exhibited a small interstitial deletion within band 7q22 (Fig 1B). This region, which is approximately 2 to 3 Mb in size based on the YAC contig map, contains the genes encoding PLANH1 and CUTL1. To detect possible submicroscopic deletions, we subsequently screened 25 CML cases with normal chromosomes 7 on G-banding analysis by FISH with YAC clones HSC7E737 and HSC7E791 from the critical region. No additional deletions of this genomic region were detected in these cases.
MDS/AML.In the 17 MDS/AML cases that exhibited 7q22 deletions, we identified a commonly deleted segment extending from YAC clone HSC7E329 in band 7q22 to clone HSC7E1289 in band 7q32 (Fig 2). The two different commonly deleted regions, which were defined by the CML and MDS/AML cases, are separated by a 380-kb genomic fragment in 7q22 recognized by YAC clone HSC7E329. The estimated size of the region deleted in all MDS/AML cases is approximately 20 Mb and contains the genes encoding PRKAR2B, DRA, CAPZA2, and MET. As shown in Fig 2, there is marked heterogeneity of the proximal deletion breakpoints; within bands 7q21-q22, the breakpoints were scattered along a large region extending from the genomic segments recognized by clone HSC7E590 (case no. 15) to clone HSC7E316 (case no. 17). In 7 cases, the deletion breakpoints were even proximal to band 7q21.1. In all but 1 case (no. 10), deletions extended distal to the genomic region identified by clone HSC7E1289 in band 7q32. Only the leukemic cells of case no. 10 showed two hybridization signals with clone HSC7E1289 and, thus, defined the distal border of the critical region of the MDS/AML cases. In 9 of the 17 deletion cases, we found loss of the telomeric sequences (detected by clone HSC7E526), suggesting that a deletion of the remaining long arm of chromosome 7 had occurred in these cases.
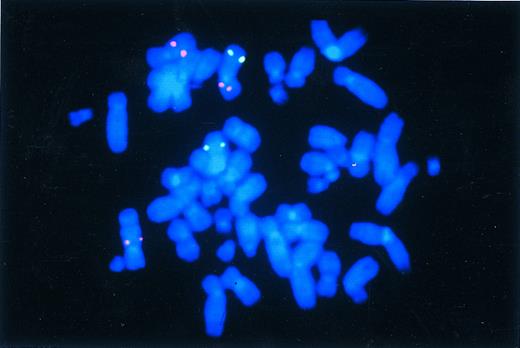
The breakpoint of the t(3; 7)(p13; q22), which was present in unstimulated bone marrow and PHA-stimulated blood in an MDS case (no. 6), was localized to the 400-kb genomic segment recognized by YAC clone HSC7E572 (Figs 1D and 3); this segment was contained in the critical region of the MDS/AML cases. The second translocation breakpoint, which we identified in 1 of the AML cases (no. 19) by G-banding analysis, was localized between clones HSC7E481 and HSC7E502. This genomic segment is located in band 7q21.3 and is thus proximal to the two commonly deleted regions.
Identification of the t(3; 7)(p13; q22) in case no. 6 by YAC clone HSC7E572 (Cy3). Note the three red signals that originate from the normal chromosome 7 (upper left), the derivative 7 (lower left), and the derivative 3 chromosome (upper right). For the identification of chromosome 3, we cohybridized with a YAC clone (fluorescein isothiocyanate) mapping to the long arm of chromosome 3.
Identification of the t(3; 7)(p13; q22) in case no. 6 by YAC clone HSC7E572 (Cy3). Note the three red signals that originate from the normal chromosome 7 (upper left), the derivative 7 (lower left), and the derivative 3 chromosome (upper right). For the identification of chromosome 3, we cohybridized with a YAC clone (fluorescein isothiocyanate) mapping to the long arm of chromosome 3.
DISCUSSION
Using FISH with well-mapped YAC clones, we identified distinct critical genomic regions in 21 cases with various myeloid disorders exhibiting deletions or reciprocal translocations of chromosome band 7q22. In the 19 deletion cases, we delineated two commonly deleted segments that were defined by the CML and MDS/AML cases, respectively.
In the 2 cases of chronic-phase CML, we delineated a narrow genomic segment in band 7q22 that was deleted in both cases. This region was defined by three overlapping YAC clones that recognize a genomic fragment of approximately 2 to 3 Mb containing the genes encoding PLANH1 and CUTL1. The second commonly deleted region was defined by the MDS/AML cases and was localized distally to the CML region. This genomic region of approximately 20 Mb encompassed the terminal part of band 7q22 and the entire band 7q31. In contrast to the study by Kere et al,10 we did not observe a narrow breakpoint cluster region of the proximal deletion breakpoints in our MDS/AML cases. The proximal breakpoints scattered along a large region extending from band 7q11 (data not shown) to band 7q22. The distal boundary of the commonly deleted segment in band 7q32 was defined by a single case (no. 10). It is likely that both commonly deleted regions, which we identified in our study, are contained within the deleted segments that were described in previous deletion mapping studies in myeloid disorders.10-12 In these studies, which used RFLP and quantitative Southern blot analyses, the proximal breakpoints were localized between the genes for EPO and PLANH1 at 7q22 in 7 patients and between the genes for elastin and collagen 1 α 2 (COL1A2) at 7q11-q21 in another 2 patients. However, the distal boundaries of the deletions were less well characterized. The proto-oncogene MET at band 7q31 was deleted in all 9 cases, whereas the gene encoding the T-cell receptor β at band 7q35 was retained in 4 of the 9 cases.11 12
Using FISH with a panel of mapped YAC clones from 7q, Le Beau et al9 recently delineated a 2- to 3-Mb commonly deleted genomic segment in band 7q22 in 15 patients with myeloid or lymphoid malignancies. This segment approximately corresponds to the genomic region identified by clones HSC7E441 through HSC7E572 that is contained within the critical region of the MDS/AML cases in our study. DNA markers common to both regions include D7S658, D7S796, D7S818, D7S2504, D7S2509, and D7S2494. Interestingly, the breakpoint of the reciprocal translocation t(3; 7)(p13; q22), which we identified in case no. 6, not only maps to the critical segment of our MDS/AML cases but also to the smallest overlapping deleted segment identified in the study by Le Beau et al.9 In this case of MDS, the translocation was present in unstimulated bone marrow and PHA-stimulated blood, indicating that the rearrangement had occurred either in an early hematopoietic precursor cell differentiating to the myeloid or lymphoid lineage or in the germline and then likely predisposed the patient to the development of MDS. This translocation breakpoint may point to a novel gene of pathogenic significance in myeloid leukemias. Translocations commonly lead to the activation of proto-oncogenes or to chimeric fusion genes of oncogenic potential. However, some translocations have been shown to be accompanied by submicroscopic deletions and have led to the identification of a genomic segment likely containing a novel tumor suppressor locus.26 By analogy, the translocation breakpoint found in our MDS case may inactivate a tumor suppressor gene, eg, by disruption of its primary structure, in particular because it occurs in a deletion cluster region.
Johnson et al13 reported on a narrow interval at band 7q22.1 in patients with MDS. The proximal breakpoint of a constitutional inversion inv(7)(22.1q34) associated with MDS as well as the breakpoint of a reciprocal translocation t(2; 7)(p13; q22) in an unrelated patient with MDS were found to be contained in a genomic fragment identified by YAC clone HSC7E783 (Table 1 and Fig 2). This finding suggests that this genomic region contains a specific gene(s) associated with the development of MDS. This region, which contains the gene for the cell-cycle control enzyme ASNS,27 maps proximally to the translocation breakpoint in case no. 6 and to the commonly deleted segments that were identified in our study and that of Le Beau et al9 (see Fig 2). In the same study, the breakpoints of two other balanced translocations associated with MDS were localized proximally and distally to the constitutional inversion breakpoint. In keeping with these findings, we mapped the breakpoint of another balanced translocation (case no. 19) between the segments identified by clones HSC7E481 and HSC7E502, which is proximal to the inversion breakpoint reported by Johnson et al.13
Taken together, the data suggest that there are multiple genes in bands 7q21-q22 that are involved in the pathogenesis of myeloid leukemias. Although the commonly deleted genomic regions are still large, the identification of inversion and translocation breakpoints provides a first step towards the cloning of these genes.
ACKNOWLEDGMENT
We gratefully acknowledge Drs U. Pascheberg, J. Mittermüller, H. Schmetzer, and B. Schlegelberger for providing leukemia specimens.
Supported by grants from the Deutsche Forschungsgemeinschaft (Do436/3-2), the European Community (GENE-CT 930055), and the Canadian Genome Analysis and Technology Program (CGAT).
Address reprint requests to Hartmut Döhner, MD, Medizinische Klinik and Poliklinik V, University of Heidelberg, Hospitalstraβe 3, 69115 Heidelberg, Germany.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal