Abstract
Translocations involving the human CBFA2 locus have been associated with leukemia. This gene, originally named AML1, is a human homologue of the Drosophila gene runt that controls early events in fly embryogenesis. To clarify the role of mammalian runt products in normal and leukemic hematopoiesis, we have studied their pattern of expression in mouse hematopoietic tissues in the adult and during ontogeny using an anti-runt box antiserum. In the adult bone marrow, we found expression of runt polypeptides in differentiating myeloid cells and in B lymphocytes. Within the erythroid lineage, runt expression is biphasic, clearly present in the erythroblasts of early blood islands and of the fetal liver, but absent in the adult. Biochemical analysis by Western blotting of fetal and adult hematopoietic populations shows several runt isoforms. At least one of them appears to be myeloid specific.
THE runt locus of Drosophila melanogaster controls early developmental events in segmentation, sex determination, and neurogenesis.1-3 The first mammalian runt homologue was identified in humans as the locus, on 21q22, involved in the t(8; 21) of acute myeloid leukemia.4,5 Because of this, it was initially named AML1. Three runt-like loci have been characterized both in humans and in mice.6-10 The official nomenclature (CBFA1-3, in which the acronym stands for Core Binding Factor, α subunit) reflects the recognition that runt products bind DNA with specificity for a consensus sequence (Pu/TACCPu/CA11,12), which was originally identified in viral enhancers13 and later also found in a number of cellular genes.14-17 The DNA binding domain has been shown to coincide with an approximately 120 amino acid sequence that is conserved among all known runt family members (runt box).12,18 At least the CBFA1 and CBFA2 loci give rise to multiple isoforms because of alternative splicing.6,7,19,20 Functional studies suggest that some of these may have opposing effects on the transcription of test target genes and on cell differentiation.21 22 Thus, current evidence suggests that runt products make up a complex regulatory system that controls critical events in differentiation.
The expression of CBFA genes has been investigated both in the adult and in the fetus at the mRNA level.23,24 However, only partial data are available for the hematopoietic system. In most adult vertebrates, hematopoiesis occurs largely in the bone marrow, where a limited pool of pluripotential stem cells gives rise, through sequential commitment and differentiation steps, to mature blood cells of the different lineages: erythrocytes, granulocytes, platelets (megakaryocytes), and lymphocytes.25 This adult or definitive hematopoietic phase is preceded, in ontogeny, by others in which hematopoietic stem cell activity can be found at three other sites: the yolk sac, the aorta-genital ridge-mesonephros (AGM) region, and the fetal liver.26,27 Significant differences are known to exist between the different phases.28 Recently, by generating mouse lines functionally deleted in the CBFA2 locus, a requirement has been postulated for this locus in hematopoiesis starting from the fetal liver phase.29 30 In contrast, the hematopoietic function of the yolk sac was apparently unimpaired in these lines. However, no direct evidence is available regarding the expression of runt polypeptides within hematopoietic cells. In this report, we set out to provide such evidence. In particular, we aimed to address the question of whether the characteristic leukemic phenotypes associated with translocations involving the human CBFA2 locus correlate with a restricted expression of normal runt products to specific lineages or stages of hematopoietic differentiation.
MATERIALS AND METHODS
Mice and embryos.TO mice were used as a source of embryos. Gestational age, which was measured in days post coitum (dpc), was calculated from midnight of the mating night. Pregnant mice were killed by cervical dislocation. Embryos were collected in cold phosphate-buffered saline (PBS) and immediately processed. The 7.5-day embryos were embedded without removing the decidua. Fetal livers were dissected with the help of a dissection microscope. Normal adult tissues were obtained from TO or CD1 mice. Bone marrow was obtained by flushing the femoral and tibial shafts with cold PBS.
Expression of recombinant proteins.Constructs encoding AML1,4 an AML1/MTG8 in-frame fusion,31 and an AML1/MTG8 out-of-frame fusion32 were assembled in eukaryotic expression vectors pBEH33 or pBABE Neo34 from genomic and cDNA fragments and verified by sequencing (F. Calabi, unpublished data). COS-7 cells35 were transfected by the diethyl aminoethyl-dextran/chloroquine procedure.36 Cells were harvested 48 hours after transfection.
A cDNA fragment encoding amino acids 51 to 128 of human AML14 was joined in frame to the glutathione-S-transferase (GST) coding sequence in the expression vector pGEX-2T (Pharmacia LKB, Uppsala, Sweden). Escherichia coli TG1 cells harboring the construct were grown in SOB36/ampicillin medium to an OD550 of 0.5 before the addition of isopropyl-β-D-thiogalactoside (IPTG; 1 mmol/L final concentration). After a further 4.5 hours, cells were harvested and lysed essentially as described.37 A significant fraction of the recombinant GST/runt box polypeptide was found to be soluble in the detergent lysate and directly purified by binding to glutathione-agarose (Sigma, Poole, Dorset, UK) and elution with 5 mmol/L reduced glutathione (GSH) in 0.1 mol/L sodium phosphate buffer, pH 8.
Preparation of the anti-runt box antiserum.A New Zealand rabbit was immunized with approximately 250 μg recombinant GST/runt box polypeptide by subcutaneous injection in complete Freund's adjuvant. Two further injections were administered in incomplete Freund's adjuvant at 10-day intervals. After 5 months of rest, the rabbit was boosted by a subcutaneous injection of approximately 10 μg purified runt box fragment. This was prepared from the affinity-purified GST/runt box polypeptide by digestion with thrombin (Sigma), followed by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The antiserum obtained from the boosted rabbit was first depleted of antibodies reactive with GST and bacterial antigens by passing it through an Affi-Gel 10 column (Bio-Rad, Richmond, CA) coupled to total cellular proteins (70 OD280 U/mL gel) from an IPTG-induced bacterial culture harboring the pGEX-2T vector. Depletion was verified by Western blotting. The anti-runt box antibodies were then further purified by affinity chromatography on an Affi-Gel 10 column coupled to affinity-purified GST/runt box polypeptide (4 mg/mL gel). Bound antibodies were eluted first with 0.1 mol/L glycine, 10% dioxan, pH 2.5, followed by 0.1 mol/L diethylamine, pH 11.5.38 Acid- and alkali-eluted fractions were dialyzed against PBS, pH 7.2; pooled; and concentrated on Centricon 30 (Amicon, Beverly, MA).
Antibodies.The following rat monoclonal antibodies (MoAbs) were used: 3C1, anti–c-kit, or CD11739 (Pharmingen, San Diego, CA); RB6-8C5, anti–Gr-140 (Pharmingen); M1/70, anti–Mac-1, or CD11b41 (Serotec, Oxford, UK); TER-119, anti-mouse erythroid cells42 (Pharmingen); and RA3-6B2, anti-CD45R, or B22043 (Sigma). A fluorescein isothiocyanate (FITC)-conjugated polyclonal rabbit anti-mouse serum (Dako, Glostrup, Denmark) was used to identify B cells.
Mouse bone marrow cell separation.The separations were performed using a VarioMacs System (Miltenyi Biotec, Bergisch-Gladback, Germany) following the manufacturer's instructions. Briefly, cells (up to 5 × 108/mL, in PBS, pH 7.2, containing 1% bovine serum albumin, 5 mmol/L EDTA, and 0.01% azide) were labeled with the rat primary antibody (TER-119 or FITC-coupled Gr-1 and CD45R, 0.5 to 1 μg/106 cells) for 30 minutes on ice, washed once, and incubated with goat anti-rat Ig coupled to Microbeads for 20 minutes at 4°C. The cells were then passed over an A2 column placed in a VarioMACS separator. Purity of the sorted cells was checked using a FACScan (Becton Dickinson, Mountain View, CA). In the case of TER-119, eluted cells were stained with FITC-conjugated mouse anti-rat Ig (Jackson, West Grove, PA) before FACS analysis. After a single passage through the magnetic field, Gr-1 and CD45R positively selected populations were usually found to be about 90% pure. However, TER-119 cells were only enriched to 50% to 60% because of cell clumping occurring on the column and causing nonspecific trapping. Therefore, in most cases, the enriched TER-119 population was dispersed by pipetting as carefully as possible and reapplied to the column. The final purity was 60% to 75%.
Immunohistochemistry.Cytospin preparations were fixed for 5 minutes in 4% paraformaldehyde in PBS, pH 7.2; washed for 2 minutes in PBS/0.5% Triton X-100; and incubated in PBS/10% goat serum/0.1% azide (blocking solution). They were then sequentially incubated with (1) anti-runt box antiserum (≈10 μg/mL) in blocking solution for 1 hour to overnight at room temperature, (2) biotinylated goat anti-rabbit antiserum (Vector, Burlingame, CA) for at least 1 hour, and (3) FITC-conjugated goat anti-biotin antiserum (Vector) or with R-Phycoerythrin-ExtraAvidin (Sigma).
Double immunolabeling on single-cell suspensions was performed as follows. After 5 minutes of preincubation on ice in blocking solution containing 0.02% azide, the anti-cell surface antigen rat MoAb was added (2 μg/106 cells) and incubated for 20 minutes. Cells were washed twice, resuspended in blocking solution, and cytospun. The slides were processed as above, adding an FITC-conjugated mouse anti-rat serum (Jackson) to samples treated with unlabeled TER-119.
For competition experiments, the purified anti-runt box antiserum was preincubated for at least 2 hours at 4°C with extracts from bacteria expressing either the GST/runt box or the GST proteins. The amount of GST/runt box extract required for complete inhibition of the anti-runt box antiserum at the working dilution was determined empirically by titration. Approximately 50 μL of antiserum, ie, the amount generally used per one slide, could usually be completely inhibited by the extract from 1 mL of bacterial culture. In every experiment, parallel samples were stained with rabbit preimmune serum as a negative control.
Slides were mounted in an antifade medium (Vector) and examined using a Zeiss Axioskop fluorescence microscope (Carl Zeiss, Jena, Germany). Images were captured by a Photometrics CCD camera (Photometrics, Tucson, AZ) coupled to a Macintosh Quadra computer (Apple Computers, Cupertino, CA). Image analysis was performed using Smartcapture Software (Digital Scientific, Cambridge, UK).
Paraffin sections were prepared for staining according to published procedures44 with minor modifications. Briefly, whole embryos and dissected organs were frozen by immersion in iso-pentane precooled to freezing point, transferred to precooled ethanol (−70°C), and left for 3 days. After this, cold ethanol was replaced every 2 days (usually 3 times) to complete dehydration. Specimens were subsequently equilibrated to room temperature and embedded in paraffin wax. Microtome sections (8-μm thick) were dewaxed in xylene (3 changes of 2 minutes each), rehydrated in graded ethanol baths, incubated in blocking solution, and processed as described above, except that the fluorescent reagent was substituted with preformed streptavidin-biotinylated horseradish peroxidase complex (Dako). Bound peroxidase was detected using di-amino-benzidine (DAB)/NiCl2 as substrate.
Western blotting.Cells were lysed in Laemmli sample buffer38 and sonicated to shear DNA; aliquots (107 cells) were fractionated by SDS-PAGE on a 9% to 18% gradient gel and blotted onto a nitrocellulose filter (Schleicher and Schüell, Dassel, Germany). The filter was blocked with 2.5% nonfat, dry milk powder in PBS before incubation for 3 hours with purified anti-runt box antiserum diluted in PBS containing 2% bovine serum albumin. After washing in PBS/0.5% Tween 20, the blot was developed with 1 μCi/10 mL 125I-protein A (>30 mCi/mg; Amersham, Buckinghamshire, UK) in PBS/0.5% Tween 20.
RESULTS
Production of an antiserum against runt box polypeptides.A polypeptide corresponding to amino acids 51 to 128 in the predicted AML1 sequence4 (ie, the first 78 amino acids of the runt box) was expressed in E coli as a fusion to GST. After affinity purification on glutathione-agarose, it was used to immunize a rabbit. Initial testing of the raw antiserum showed reactivity against cellular components that was not blocked by the GST/runt box polypeptide. The antiserum was then affinity purified as described in Materials and Methods to yield an operationally specific reagent, as tested on Western blots of expressing bacterial lysates (data not shown). As a further stringent specificity criterion, in most of our immunostaining experiments we have made use, as a negative control, of purified antiserum blocked by preincubation with a bacterial lysate expressing the GST/runt box polypeptide. This was compared with the signal given by the same antiserum preincubated with a bacterial lysate expressing a comparable amount of GST alone. We are therefore highly confident of the specificity of the staining observed for runt polypeptides.
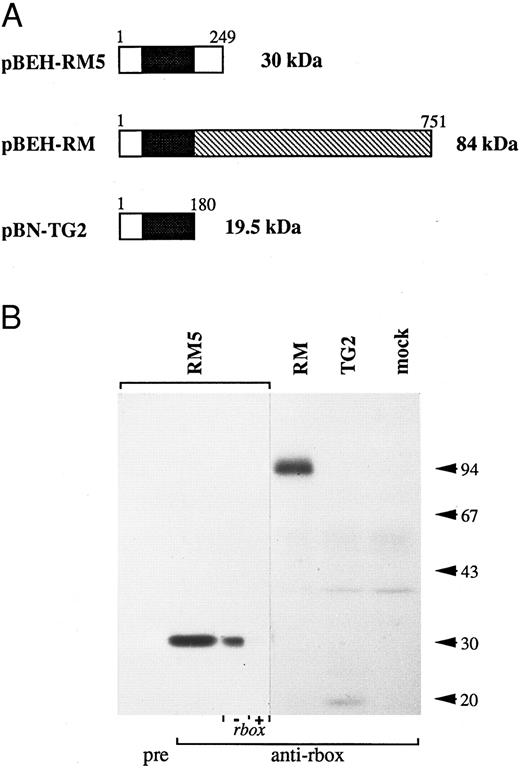
The reactivity of the anti-runt box antiserum was verified by Western blotting analysis of a panel of COS-7 cells expressing three different runt box-containing polypeptides (Fig 1): a 30-kD polypeptide corresponding to AML14 and encoded by the pBEH-RM5 plasmid; an 84-kD polypeptide corresponding to an in-frame AML1/MTG8 fusion31 and encoded by the pBEH-RM plasmid; and a 19.5-kD polypeptide corresponding to an out-of-frame AML1/MTG8 fusion32 and encoded by the pBN-TG2 plasmid. In each case, the major detected species migrated at or very close to the predicted molecular weight.
Reactivity of the anti-runt box antiserum against runt box polypeptides. (A) Schematic diagram of the different polypeptides tested. The runt box is in black; the chromosome 8 sequence in pBEH/RM is indicated by the hatched box; a short sequence of chromosome 8 origin is also present at the 3′ end of the runt box in plamid pBN-TG2. Numbers refer to amino acids. The predicted molecular weight is on the right. (B) Western blotting analysis of COS-7 cells transiently transfected with plasmids encoding the described runt box polypeptides. “pre” and “anti-rbox” refer to the use of preimmune serum or of affinity-purified anti-runt box antiserum, respectively. As a further specificity check, the pBEH-RM5 lane was split into two and probed with purified anti-runt box antiserum either mock-depleted (by preincubation with matrix-bound GST, “− rbox”) or depleted of anti-GST/runt box reactivity (by preincubation with matrix-bound GST/runt box, “+ rbox”). mock, mock-transfected COS cells. The three leftmost lanes were exposed for 18 hours, the others for 3 days.
Reactivity of the anti-runt box antiserum against runt box polypeptides. (A) Schematic diagram of the different polypeptides tested. The runt box is in black; the chromosome 8 sequence in pBEH/RM is indicated by the hatched box; a short sequence of chromosome 8 origin is also present at the 3′ end of the runt box in plamid pBN-TG2. Numbers refer to amino acids. The predicted molecular weight is on the right. (B) Western blotting analysis of COS-7 cells transiently transfected with plasmids encoding the described runt box polypeptides. “pre” and “anti-rbox” refer to the use of preimmune serum or of affinity-purified anti-runt box antiserum, respectively. As a further specificity check, the pBEH-RM5 lane was split into two and probed with purified anti-runt box antiserum either mock-depleted (by preincubation with matrix-bound GST, “− rbox”) or depleted of anti-GST/runt box reactivity (by preincubation with matrix-bound GST/runt box, “+ rbox”). mock, mock-transfected COS cells. The three leftmost lanes were exposed for 18 hours, the others for 3 days.
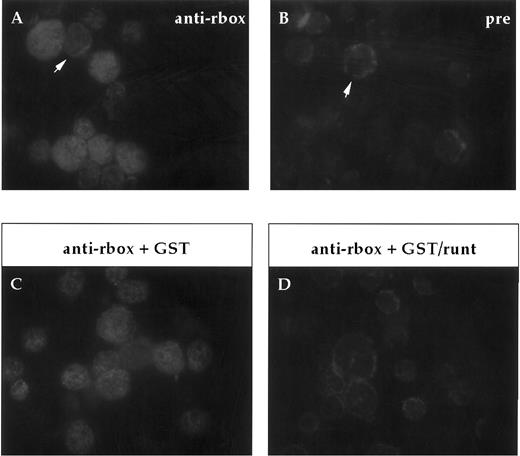
Expression of runt box polypeptides in adult bone marrow.In our analysis of the expression of runt box polypeptides in hematopoietic cells, we focused on the mouse to be able to study more easily both the adult and the embryonic/fetal stages. First of all, expression in adult bone marrow was investigated on cytospin preparations. Most cells were found positive for runt (Fig 2), ranging from large blasts to maturing granulocytes. A clear positive signal was detected in the nucleus. It was somewhat stronger and more homogenous in the larger, more immature cells.
Immunofluorescent labeling of adult bone marrow cells with anti-runt box antiserum. (A and B) Purified anti-runt box antiserum (anti-rbox) stains the nuclei of bone marrow cells with variable intensity (A). Occasional cells show weak cytoplasmic staining (arrow), but this is also visible with the preimmune serum (pre) (B, arrow). (C and D) Staining with anti-runt box antiserum preincubated with a bacterial extract containing either GST (anti-rbox + GST; C) or GST/runt (anti-rbox + GST/runt; D). The staining is abolished only in the latter sample.
Immunofluorescent labeling of adult bone marrow cells with anti-runt box antiserum. (A and B) Purified anti-runt box antiserum (anti-rbox) stains the nuclei of bone marrow cells with variable intensity (A). Occasional cells show weak cytoplasmic staining (arrow), but this is also visible with the preimmune serum (pre) (B, arrow). (C and D) Staining with anti-runt box antiserum preincubated with a bacterial extract containing either GST (anti-rbox + GST; C) or GST/runt (anti-rbox + GST/runt; D). The staining is abolished only in the latter sample.
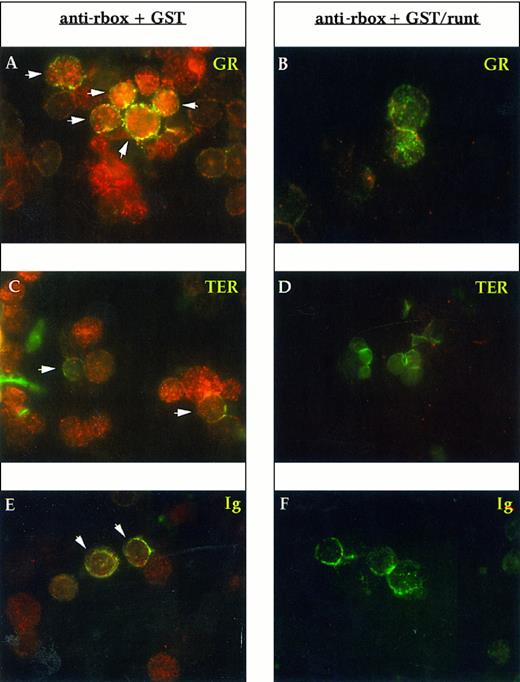
To determine the lineage of runt-expressing cells, we performed double staining with lineage-specific markers. Gr-1 and Mac-1 were used for myelomonocytic elements, the former being specific for the granulocytic lineage, whereas the latter is shared by granulocytes and monocytes; TER-119 for erythroid cells; and Ig for B cells. Essentially all Gr-1+ cells express runt (Fig 3A). In preparations developed with a peroxidase-conjugated anti-rabbit antiserum and counterstained with Mayer hemalum, we observed that the smaller cells with the doughnut-shaped nucleus characteristic of mature granulocytes appear to be less positive than the larger, more immature cells (data not shown). Erythroid cells, which were identified by the TER-119 antibody, do not show a clear reactivity; some dubious spots can be detected in only about 30% to 50% of the cells (Fig 3C). B-lineage cells (Ig+) are uniformly positive (Fig 3E).
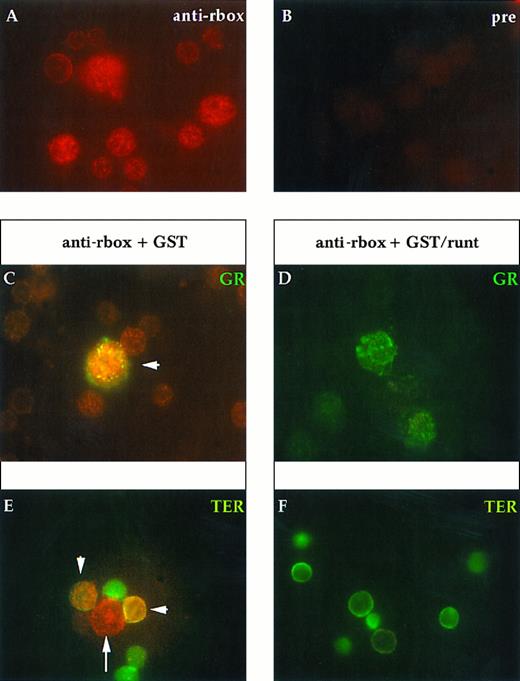
Immunofluorescent labeling of adult mouse bone marrow cytospin preparations with anti-runt box antiserum in combination with MoAbs specific for granulocytes (anti-Gr-1; A and B) and erythroid cells (TER-119; C and D) or with an antiserum against B lymphocytes (anti-mouse Ig; E and F ). The anti-runt box reactivity appears as red-orange (R-phycoerythrin); reactivity with lineage-specific antibodies appears in green (FITC). The anti-runt box was competed with GST (anti-rbox + GST; A, C, and E) or with GST/runt (anti-rbox + GST/runt; B, D, and F ). Double-positive cells in (A), (C), and (E) are indicated by arrows.
Immunofluorescent labeling of adult mouse bone marrow cytospin preparations with anti-runt box antiserum in combination with MoAbs specific for granulocytes (anti-Gr-1; A and B) and erythroid cells (TER-119; C and D) or with an antiserum against B lymphocytes (anti-mouse Ig; E and F ). The anti-runt box reactivity appears as red-orange (R-phycoerythrin); reactivity with lineage-specific antibodies appears in green (FITC). The anti-runt box was competed with GST (anti-rbox + GST; A, C, and E) or with GST/runt (anti-rbox + GST/runt; B, D, and F ). Double-positive cells in (A), (C), and (E) are indicated by arrows.
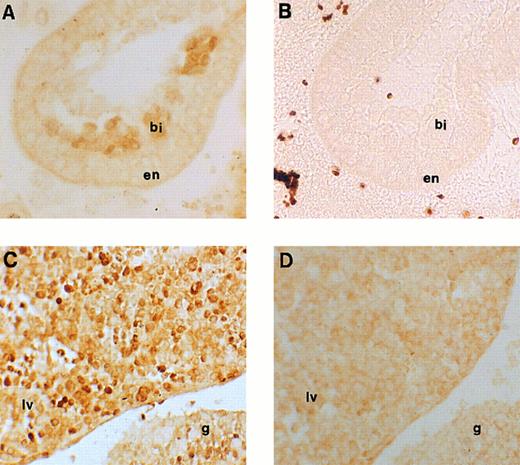
Immunohistochemical labeling of early erythroid cells. (A) Section through the 7.5-dpc yolk sac showing staining for runt box polypeptides in the developing blood islands (original magnification × 20). (B) Negative control for (A), ie, an adjacent section stained with anti-runt box antiserum after preincubation with GST/runt polypeptide. Given the lack of stain, a phase contrast image is shown for better definition (original magnification × 20). bi, blood island; en, extraembryonic endoderm. (C) Section through a 11.5-dpc mouse embryo, showing staining for runt box polypeptides in the fetal liver (lv) and in the urogenital ridge (g). The neighboring tissues are negative (original magnification × 20). (D) Same as (C), but preimmune control (original magnification × 20).
Immunohistochemical labeling of early erythroid cells. (A) Section through the 7.5-dpc yolk sac showing staining for runt box polypeptides in the developing blood islands (original magnification × 20). (B) Negative control for (A), ie, an adjacent section stained with anti-runt box antiserum after preincubation with GST/runt polypeptide. Given the lack of stain, a phase contrast image is shown for better definition (original magnification × 20). bi, blood island; en, extraembryonic endoderm. (C) Section through a 11.5-dpc mouse embryo, showing staining for runt box polypeptides in the fetal liver (lv) and in the urogenital ridge (g). The neighboring tissues are negative (original magnification × 20). (D) Same as (C), but preimmune control (original magnification × 20).
The c-kit receptor is helpful in defining an earlier differentiation stage than lineage markers, because approximately 50% of the positive cells are negative for lineage markers. About 70% of the c-kit–bearing cells are runt-positive (data not shown), suggesting that runt expression is also found in some precursor cells, ie, before terminal differentiation.
Because monocytes represent only a minor fraction of bone marrow cells (≈2%), expression of runt in the monocytic lineage was investigated on a nearly pure population of residential peritoneal macrophages (data not shown) and found to overlap completely with that of Mac-1 (corresponding to 70% of the cells).
The megakaryocytic lineage was not studied in detail. However, the large hyperdiploid megakaryocytes, which are easily recognizable in the bone marrow, are runt-negative (data not shown).
Expression of runt box polypeptides in embryonic hematopoiesis.We next moved to analyze runt expression in early hematopoiesis. The first time point examined was at 7.5 dpc. At this time, blood islands begin to form in the mesoderm of the yolk sac and erythroblasts can be seen.45 Specific nuclear staining was observed with the anti-runt box antiserum in most of the cells within these structures (Fig 4A and B).
Immunofluorescent labeling of 16.5-dpc fetal liver cells with anti-runt box antiserum either alone (A and B) or in combination with MoAbs specific for granulocytes (anti-Gr-1; C and D) or erythroid cells (TER-119; E and F ). The anti-runt box reactivity appears as red-orange stain (R-phycoerythrin); reactivity with lineage-specific antibodies appears in green (FITC). (A) Anti-runt box staining. (B) Preimmune rabbit serum control. Competitions with GST or GST/runt (C through F ) were performed as described in the legend to Fig 2. In (C), the arrow indicates a large blast of granulocytic lineage (green staining) that was strongly labeled by the anti-runt box (red-orange); this last staining is prevented by preincubation with GST/runt. In (E), the double-labeled cells (arrowhead) show expression of runt-box polypeptides in the erythroid lineage, albeit at much lower levels than in myeloid blasts (long arrow). Even in these cells, specificity of the anti-runt box stain is shown by competition with GST/runt (F ).
Immunofluorescent labeling of 16.5-dpc fetal liver cells with anti-runt box antiserum either alone (A and B) or in combination with MoAbs specific for granulocytes (anti-Gr-1; C and D) or erythroid cells (TER-119; E and F ). The anti-runt box reactivity appears as red-orange stain (R-phycoerythrin); reactivity with lineage-specific antibodies appears in green (FITC). (A) Anti-runt box staining. (B) Preimmune rabbit serum control. Competitions with GST or GST/runt (C through F ) were performed as described in the legend to Fig 2. In (C), the arrow indicates a large blast of granulocytic lineage (green staining) that was strongly labeled by the anti-runt box (red-orange); this last staining is prevented by preincubation with GST/runt. In (E), the double-labeled cells (arrowhead) show expression of runt-box polypeptides in the erythroid lineage, albeit at much lower levels than in myeloid blasts (long arrow). Even in these cells, specificity of the anti-runt box stain is shown by competition with GST/runt (F ).
At 11.5 dpc, the yolk sac is near the end of its erythropoietic function, although it is still the source of all circulating erythrocytes. In contrast, the liver is the expanding hematopoietic organ and is constituted by immature, fast growing cells. These cells are almost exclusively erythroid,25 although for the most part lacking the erythroid-specific marker TER-119, whereas they are strongly positive for c-kit.46 On wax sections, the anti-runt box antiserum stained about half of these cells (Fig 4C and D) with variable intensity. Cytospin preparations were also made from the liver of 12.5-dpc mice to study in greater detail the nature of the positive cells and the localization of the runt box polypeptides. Both early erythroblasts and nucleated primitive erythrocytes could be identified (Fig 5A through D). The former, which are easily recognizable by their nuclear shape and high nucleus/cytoplasmic ratio, showed a diffuse nuclear staining. However, in the latter cells, which are characterized by their round nuclei and lower nucleus/cytoplasmic ratio, staining was visible only at the nucleus/cytoplasmic border (Fig 5A, arrowheads). An identical staining pattern can be observed in the nucleated primitive erythrocytes that make up the majority of circulating blood cells at this stage (Fig 5E). It is unlikely that this pattern is due to a fixation or staining artefact, because it was apparent even with a different fixative (ie, cold acetone for 10 minutes) and with a peroxidase-conjugated anti-rabbit reagent (data not shown).
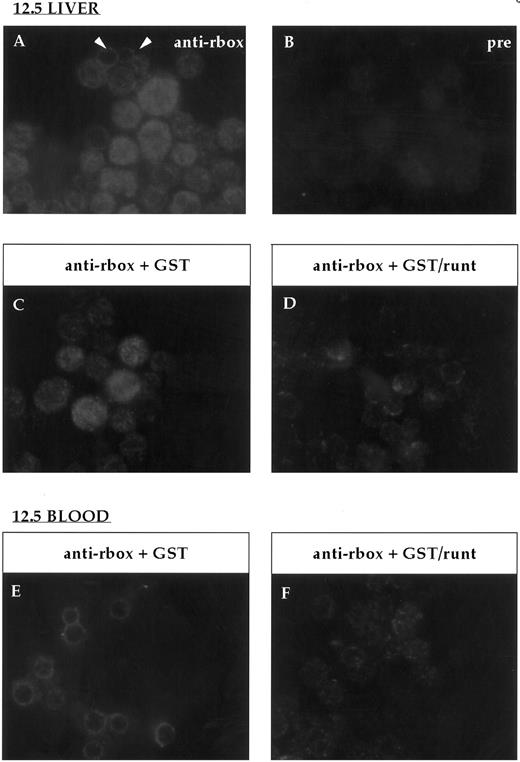
Immunofluorescent labeling of cytospin preparations from 12.5-dpc liver and blood stained with the anti-runt box antiserum. (A) Anti-runt box staining of liver cells; two primitive erythrocytes labeled at the nucleus-cytoplasmic border are indicated (arrows). (B) Staining with preimmune serum. Competition experiments were performed on 12.5-dpc liver (C and D) and blood (E and F ) as for bone marrow (Fig 2). In both tissues, the staining persisted after preincubation with GST (C and E), but was abolished after preincubation with GST/runt (D and F ).
Immunofluorescent labeling of cytospin preparations from 12.5-dpc liver and blood stained with the anti-runt box antiserum. (A) Anti-runt box staining of liver cells; two primitive erythrocytes labeled at the nucleus-cytoplasmic border are indicated (arrows). (B) Staining with preimmune serum. Competition experiments were performed on 12.5-dpc liver (C and D) and blood (E and F ) as for bone marrow (Fig 2). In both tissues, the staining persisted after preincubation with GST (C and E), but was abolished after preincubation with GST/runt (D and F ).
At 16.5 days, the liver contains erythroid cells of all differentiation stages, displaying almost all the characteristics of the adult bone marrow erythroid cells. However, populations differentiating toward the other lineages can be shown by both morphologic analysis and staining with lineage-specific antibodies. Overall, more than one third of the cells express nuclear runt box polypeptides (Fig 6A). The perinuclear localization observed in primitive erythrocytes at the earlier time point is no longer visible. Double staining analyses are shown in Fig 6C through F. All the maturing (Gr-1+) granulocytic elements (about 5% in our samples) were strongly stained for runt. About 30% of the TER-119+ cells were stained by the anti-runt box antiserum, showing that runt expression is maintained in some erythroid cells at this stage.
The proportion of runt-expressing cells within the c-kit+ population was lower in 16.5-dpc fetal liver (40%; data not shown) than in adult bone marrow (70%; see above). This correlates with a higher proportion of immature precursors within the c-kit+ pool of the former tissue46 and suggests that runt is expressed by the more mature c-kit+ cells.
Of the Mac-1+ cells (6% of the total), approximately 60% expressed runt (data not shown). Because Gr-1+ granulocytes make up a similar proportion of the Mac-1+ population and they are all runt-positive, it is likely that the Mac-1+/runt− cells are Gr-1−, ie, they must be monocytes.
Immunochemical analysis of runt box polypeptides. Western blotting analysis on adult bone marrow cell lysates showed a number of runt box polypeptides (Fig 7A). The most abundant component migrated as a doublet at an apparent molecular weight of 53/55 kD, and minor ones at 43/45, 40/41, and 34 kD. We therefore designate the runt products as p53/55-runt, p43/45-runt, p40/41-runt, and p34-runt. Bands at 37 and 30 kD were recognized by the preimmune serum (Fig 7A, asterisks) and were not competed by the recombinant GST/runt box polypeptide (data not shown). Therefore, they must be regarded as nonspecific. In some, but not all experiments, we could also detect an additional specific band at approximately 27 kD. This was only found in some TO mice and never in CD1, and may suggest a certain variability in the pattern of runt expression. In general, the relative abundance of the various species appeared to vary to a certain extent between different experiments (compare the bone marrow lanes in Fig 7A and C).
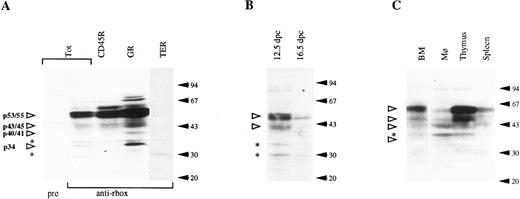
Western blotting analysis of hematopoietic cell populations with the anti-runt box antiserum. (A) Lysates from total bone marrow cells (Tot) or from bone marrow subpopulations enriched for expression of CD45R, Gr-1 (GR), and TER-119 (TER). pre, preimmune serum; anti-rbox, anti-runt box antiserum. (B) Lysates from 12.5- and 16.5-dpc liver cells. (C) Lysates from peritoneal macrophages (Mφ ), thymus, and spleen cells. The total bone marrow lysate (BM) is shown for reference. In all panels, each lane corresponds to 107 cells. Specifically detected polypeptides are indicated by the open arrowheads on the left, with the estimated molecular weight given. Asterisks mark nonspecific bands (see text). The positions of the molecular weight markers are indicated on the right (solid arrows).
Western blotting analysis of hematopoietic cell populations with the anti-runt box antiserum. (A) Lysates from total bone marrow cells (Tot) or from bone marrow subpopulations enriched for expression of CD45R, Gr-1 (GR), and TER-119 (TER). pre, preimmune serum; anti-rbox, anti-runt box antiserum. (B) Lysates from 12.5- and 16.5-dpc liver cells. (C) Lysates from peritoneal macrophages (Mφ ), thymus, and spleen cells. The total bone marrow lysate (BM) is shown for reference. In all panels, each lane corresponds to 107 cells. Specifically detected polypeptides are indicated by the open arrowheads on the left, with the estimated molecular weight given. Asterisks mark nonspecific bands (see text). The positions of the molecular weight markers are indicated on the right (solid arrows).
To exclude that the apparent complexity of runt polypeptides shown on Western blotting analysis was due to partial proteolysis, in some experiments transfected COS-7 cells expressing the AML1a polypeptide were mixed with bone marrow cells before lysis and electrophoresis. No decrease in the amount or size of the COS cell-derived AML1a polypeptide was detected in comparison with unmixed samples (data not shown).
To investigate whether there is lineage-restricted expression of individual runt products, Western blotting was also performed on purified bone marrow subpopulations expressing CD45R, Gr-1, or TER-119 markers. B lymphocytes and granulocytes account for most, if not all, of the runt-box polypeptides in the whole bone marrow (Fig 7A). All of the described runt products appear to be expressed in both cell types, with the exception of p34-runt, which is significantly enriched in the myeloid cells, while totally absent in B lymphocytes. Furthermore, the Gr-1+ population shows a higher average content of p53/55-runt than do total marrow or B cells, and only the smaller component of the p40/41-runt doublet is visible.
In Fig 7A, other slower migrating species seem also to be enriched in either myeloid or B cells. However, these results were not reproducible. Consistent with the immunohistochemistry data, no specific runt signal can be seen in the adult TER-119+ (erythroid) population.
Western blotting analysis was also performed on fetal liver cell lysates from 12.5- and 16.5-dpc embryos (Fig 7B). The pattern is essentially similar to that of whole adult bone marrow. However, the intensity of the bands decreases with developmental age. The presence of runt box polypeptides in erythroid (TER-119+) populations from 16.5 fetal liver, but not from adult bone marrow, is unlikely to be due solely to a different cellular complexity. Morphologic analysis by May-Grünwald-Giemsa staining showed that the samples had a similar composition in differentiated elements, whereas the difference in the relative content of nucleated cells (56% in the 16.5-day fetal liver sample against 22% in the adult bone marrow sample) was less than 3 times.
We also investigated the runt box polypeptides expressed at other adult hematopoietic sites, ie, thymus, spleen, and peritoneal macrophages (Fig 7C). All of the examined samples express p53/55-runt, p43/45-runt, and p40/41-runt, although their relative abundance varies. For example, p40/41-runt is the prevalent species in peritoneal macrophages, although it could not be clearly detected in the thymus. p34-runt was not observed in any of these tissues.
DISCUSSION
The involvement of a human runt homologue (CBFA2, at 21q22) in a number of leukemia-associated chromosome translocations47-49 suggests that it plays a crucial role in hematopoiesis. Consistent with this finding, runt products have been shown to be able to regulate genes whose expression is characteristic of differentiated blood cells,14-17 to modulate the ability of hematopoietic cell lines to differentiate in vitro,20 and to be required for normal fetal liver hematopoiesis.29 30 However, a satisfactory definition of the role of mammalian runt genes in hematopoiesis requires a detailed analysis of their pattern of expression within the different lineages and at different stages. This is of particular interest in view of the distinctive leukemic phenotypes associated with different CBFA2 rearrangements. Furthermore, although a number of runt isoforms have been predicted on the basis of cDNA cloning, their direct biochemical analysis has so far lagged behind. We have approached these issues by developing an antiserum specific for runt polypeptides. This antiserum was raised against the NH2 -terminal half of the human runt box encoded by the CBFA2 locus. However, given the degree of both inter-species and inter-isotype sequence conservation, it must be expected to recognize all the known runt box-containing polypeptides. The antiserum has been used first, in conjunction with hematopoietic lineage/stage markers, for an analysis of expression at the single-cell level and, second, for biochemical characterization.
At the cellular level in adult mouse bone marrow, runt polypeptides are found in differentiating granulocytic and B-lymphoid cells. They are also found in other hematopoietic lineages outside the bone marrow (ie, thymocytes and peritoneal macrophages). Thus, even within the hematopoietic system, runt expression does not coincide with commitment to a single lineage.
Typical, mature granulocytes in mouse bone marrow have been reported to be negative for cbfa2 mRNA by in situ hybridization.24 The discrepancy with our results may be due to the different sensitivity of the techniques used. We have found the smaller, doughnut-shaped cells to give the weakest runt signals. Alternatively, runt polypeptides may persist in late myelocytes well after their synthesis has ceased, reflecting a slow turnover, as reported for other myeloid proteins.50
Within the erythroid lineage, runt expression undergoes two phases during ontogenesis. Whereas runt products are essentially undetectable in the adult, they are clearly expressed in the embryo. This is true not only of the yolk sac stage, but also at least of the beginning of the liver phase (11.5 dpc). However, by late fetal life (16.5 dpc) the frequency of runt-positive erythroid precursors in the liver decreases. This decrease may correlate with the different origin, during ontogenesis, of fetal liver erythroid precursors. It is currently generally accepted that sequential waves of hematopoietic activity are contributed to the fetal liver by extra-embryonic (ie, the yolk sac) and intra-embryonic (the AGM region) sources.28 The decrease in runt expression may parallel the decline in erythropoieitic activity of yolk sac progenitors, which are replaced by intra-embryonic progenitors.
Recent experiments have shown that, whereas liver hematopoiesis appears to require a functional CBFA2 locus, yolk sac erythropoiesis would not.29 30 In the light of our experimental evidence on the expression of runt polypeptides in the yolk sac, these data suggest two hypotheses. First, because our anti-runt box antiserum would cross-react with CBFA1 and CBFA3 products, it is possible that either or both latter loci, rather than CBFA2, are expressed in the yolk sac. Second, although normally expressed and presumably required in yolk sac hematopoiesis, CBFA2 products may be functionally replaced, within this system, by other gene products. Analogous examples of plasticity and functional redundancy have been described in several other developmental pathways. A number of genes have been shown to be required for yolk sac hematopoiesis, eg, GATA-1,tal1,rbtn2, and GATA-2.51-54 It will be interesting to investigate the relationships, if any, between these genes and runt.
Although runt polypeptides are clearly expressed in maturing granulocytes, B lymphocytes, and monocytes, their presence at earlier differentiation stages remains to be investigated. Our data on c-kit+ cells suggest that they are expressed before overt lineage differentiation. Furthermore, we have preliminary evidence that human CD34+ cells are runt-positive (M.T.C., unpublished data).
At the biochemical level, our analysis shows that a number of runt box polypeptides are present in both primitive and definitive hematopoietic cells. We have not established the precise identity of the various species observed. However, they can be correlated with the isoforms predicted from cDNA sequences by comparison of the observed with the expected molecular weights. Thus, the p53/55-runt that is the major component in both total bone marrow and purified granulocytic and lymphoid subpopulations may correspond to a CBFA1 and/or to a CBFA2 isoform (ie, the A119 or the B16 isoform, respectively). A slightly higher molecular weight has been reported for the former expressed in COS cells.55 p40/41 runt may correspond to the CBFA2 B2 isoform6 and p34 runt may correspond to the CBFA1 A2 isoform.19 As for the p43/45 runt, it may correspond to the product of the CBFA3 locus, based on the sequence of a human cDNA.7 The three largest species appear as doublets, ie, two closely spaced bands of roughly similar abundance. This finding suggests that they share a similar type of modification (eg, phosphorylation). The variable ratio between individual species may indicate that their relative balance is dynamic and possibly correlated with different functional states.
Clearly, at least some of the complexity of runt box polypeptides in the bone marrow may be due to cellular heterogeneity. However, with one notable exception (the p34 runt, which is specific to granulocytes), a largely similar pattern was observed in more homogenous subpopulations of individual lineages, ie, relatively mature granulocytes, B lymphocytes, thymocytes, and embryonic erythroid precursors. It can be concluded that the components of this pattern (ie, p53/55 runt, p43/45 runt, and p40/41 runt) are not sufficient per se to induce lineage commitment in hematopoietic precursors. It remains possible that they play a role in this process or that they are involved in postcommitment events shared by the different lineages.
Finally, the data presented here bear on the relationship between rearrangements at the human CBFA2 locus and leukemia. Both myeloid and lymphoid leukemia have been associated with translocations involving this locus,47-49 although the two types correlate with different breakpoint positions. The phenotypes of the leukemic populations resemble either of the main cellular types expressing runt polypeptides in normal bone marrow. Thus, the immunophenotype of t(12; 21) leukemias (HLA-DR+, CD10+, and CD19+56) indicates a B-cell origin and correlates with runt expression in normal marrow B cells. The phenotype of t(8; 21) AML, which is characterized by a certain degree of maturation within the leukemic clone, correlates with CBFA expression in differentiating myelocytes. This finding suggests that leukemic transformation has a requirement for the same conditions that allow normal CBFA expression or action, rather than being due to inappropriate lineage or stage expression. It may be expected that other lineages expressing runt polypeptides are susceptible to leukemic transformation upon mutation at one of the CBFA loci. Alternatively, it is possible that the novel translocated product(s) encoded by a rearranged CBFA2 locus competes or synergizes selectively with a lineage- or stage-specific runt isoform. A prime candidate in the myeloid lineage would be the p34-runt.
ACKNOWLEDGMENT
We are very grateful to Dr A. Daga for assembling the pGEX/runt box construct, to Drs A. Fleming and A. Copp for providing time-mated mice, to Dr K. Kotowicz for help with the VarioMACS System, to Dr H. Kempski for help with the Photometrics CCD camera, to Prof P. Thorogood for encouragement, and to Prof P. Thorogood and Dr I. Vielle-Grosjean for critically reading the manuscript. We thank Prof J. Goldman and the Department of Haematology, Royal Postgraduate Medical School, for their generous hospitality during the early stages of this work.
Supported by MRC Programme Grant No. PG9311737. M.T.C. was supported by an EMBO long-term fellowship.
Address reprint requests to Franco Calabi, MD, Developmental Biology Unit, Division of Cell and Molecular Biology, Institute of Child Health, 30 Guilford St, London WC1N 1EH, UK.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal