Abstract
Leukemic cells from a subset of children with acute lymphoblastic leukemia (ALL) express lymphoid antigens of both T lineage and B lineage, but the clinical significance of this immunophenotype is unknown. We now report the first comprehensive comparison of treatment outcomes among a large cohort of children with CD2+CD19+ biphenotypic ALL (N = 77), B-lineage ALL (BL) (N = 1,631), or T-lineage ALL (TL) (N = 347) ALL who were treated on risk-adjusted Children's Cancer Group (CCG) protocols. CD2+CD19+ patients were more similar to BL than TL patients with respect to presenting features and antigen expression. The percentages of patients achieving successful induction therapy outcome were 98.7%, 97.8%, and 97.3% for CD2+CD19+, BL, and TL patients, respectively. Univariate comparisons of 4-year event-free survival (83.7%, 72.8%, 75.2% for CD2+CD19+, BL, and TL patients, respectively) achieved borderline significance (CD2+CD19+ B, P = .08; CD2+CD19+v T, P = .07). Relative hazard rate (RHR) estimates for BL and TL compared with CD2+CD19+ were 1.79 and 1.90, respectively, implying a better outcome for biphenotypic patients. However, multivariate adjusted RHRs for BL and TL compared with CD2+CD19+ were 1.43 (P = .29) and 1.16 (P = .76), respectively, suggesting a significant reduction in risk for BL or TL patients once adjustment was made for the more favorable characteristics of the CD2+CD19+ group. Thus, pediatric ALL patients treated on contemporary CCG protocols who present with CD2+CD19+ biphenotypic leukemia generally have good treatment outcomes, due in part to their favorable presenting features.
LEUKEMIC CELLS from patients with acute lymphoblastic leukemia (ALL) express a variety of surface differentiation antigens that are also found on normal lymphocyte precursors at discrete stages of maturation.1-3 Therefore, ALL cells are thought to originate from normal lymphocyte precursors arrested at early stages of either B-cell or T-cell ontogeny.1-4 Although leukemic cells from most ALL patients express either B-lineage or T-lineage differentiation antigens, leukemic cells from a small fraction of patients coexpress both CD19, a B-lineage surface antigen, and CD2, a T-lineage surface antigen.5-7 The clinical significance of coexpression of CD2 and CD19 antigens on leukemic cells from biphenotypic ALL patients is largely unknown. Therefore, we have analyzed the presenting features and treatment outcomes of CD2+CD19+ newly diagnosed pediatric ALL patients enrolled in risk-adjusted treatment protocols of the Children's Cancer Group (CCG). Our results show that children with biphenotypic CD2+CD19+ ALL are generally similar to B-lineage ALL (BL) patients and different from T-lineage ALL (TL) patients with respect to both presenting features and composite immunophenotype of their leukemia. Univariate analysis of outcome showed a more favorable event-free survival (EFS) outcome for CD2+CD19+ ALL compared with either BL or TL. However, no significant differences in EFS outcome were observed in multivariate analyses adjusting for other prognostic factors. Thus, our findings provide evidence that pediatric ALL patients who present with CD2+CD19+ biphenotypic leukemia exhibit favorable treatment outcomes on contemporary CCG protocols, and this result is due in part to their favorable presenting features.
MATERIALS AND METHODS
Patients.The study population for these analyses included patients (<21 years old) with newly diagnosed ALL enrolled between January 1, 1989 and December 31, 1993 on one of five risk-adjusted CCG treatment protocols (see below) for whom CD2 and CD19 antigen expression data were obtained. Diagnosis of ALL was based on morphological, biochemical, and immunological features of the leukemic cells, including lymphoblast morphology on Wright-Giemsa–stained bone marrow (BM) smears, positive nuclear staining for terminal deoxynucleotidyl transferase (TdT), negative staining for myeloperoxidase, and cell surface expression of two or more lymphoid differentiation antigens (see below). Standard definitions of remission, relapse, and induction failure were previously reported.8-10 Degree of organomegaly (moderate or marked enlargement) was as defined previously.11 CCG risk adjusted ALL protocols were as follows: CCG 1881 (low-risk protocol for children age 2 to 9 years and white blood cell [WBC] count <10,000/μL); CCG 1882 (high-risk protocol for patients age 1 to 9 years with WBC count ≥50,000/μL or age ≥10 years); CCG 1883 (protocol for infants <1 year of age), 1891 (intermediate-risk protocol for children age 2 to 9 years and WBC count 10,000 to 49,999/μL or age 1 year and WBC count <50,000/μL); and CCG 1901 (high-risk protocol for patients with lymphomatous features). Lymphomatous features were essentially as described by the revised criteria of Steinherz et al.11 Each protocol was approved by the both National Cancer Institute (NCI) and the Institutional Review Boards of the participating CCG-affiliated institutions. Informed consent was obtained from parents, patients, or both, as deemed appropriate, for both treatment and laboratory studies according to Department of Health and Human Services guidelines. For comparisons of presenting features, antigen expression, and therapy outcomes, patients were classified immunophenotypically as either CD2+CD19+ biphenotypic ALL, BL, or TL. Comparisons also were done according to recently published NCI risk classification criteria,12 which define patients with age 1 to 9 years and WBC count <50,000/μL as standard risk and all other patients as high risk.
Immunophenotyping.Mononuclear cell fractions comprised primarily (≥90%) of leukemic cells were isolated from pretreatment BM aspirate samples by centrifugation on Ficoll-Hypaque gradients (Sigma, St Louis, MO). Immunophenotyping was performed centrally in the CCG ALL Biology Reference Laboratory by indirect immunofluorescence and flow cytometry using monoclonal antibodies (MoAbs) reactive with the following lymphoid differentiation antigens: CD1, CD2, CD3, CD4, CD5, CD7, CD8, CD9, CD10, CD13, CD19, CD20, CD21, CD22, CD33, CD34, CD40, and CD72, as previously described.5,13 14 Antigen expression data are presented as the mean ± standard error (SE) and median percentages of leukemic cells scored positive for expression of a given antigen. The term “expression frequency” is used throughout to indicate the percentage of leukemic cells expressing a given antigen. Patients were classified as CD2+CD19+ biphenotypic ALL if ≥30% of their blasts were positive for both CD2 and CD19, and if >70% were positive for either CD2 or CD19.
Statistical methods.CD2+CD19+ ALL patients were compared with BL and TL controls for similarity of various clinical, demographic, and laboratory features using chi-square tests for homogeneity of proportions. Comparisons of antigen expression frequency used the Kruskal-Wallis nonparametric rank test.15 Most of the outcome analyses used life table methods and associated statistics. The primary endpoint examined was EFS from entry on study. EFS events included induction failure (nonresponse to therapy or death during induction), leukemic relapse at any site, death during remission, or second malignant neoplasm, whichever occurred first. Patients not experiencing an event at the time of EFS analysis were censored at the time of their last contact. Follow-up for event-free survivors ranged from 1 to 73 months (median, 32 months). Life table estimates were calculated by the Kaplan-Meier (KM) procedure, and the standard deviation of the life table estimate was obtained using Greenwood's formula.16 To indicate precision, the KM estimate of EFS and its standard deviation (SD) are provided at selected time points. An approximate 95% confidence interval can be obtained from the life table estimate ± 1.96 SDs.
Life table comparisons of EFS outcome pattern for patient groups generally used the log-rank statistic.17,18P values for life table comparisons are based on the pattern of outcome across the entire period of patient follow-up, but also may be given at specific time points for comparative purposes. P values ≤.05 are referred to as significant; P values between .05 and .10 are referred to as having borderline significance. Multivariate analysis of the effect of biphenotypic ALL status on EFS outcome was performed using the Cox proportional hazards model to adjust for patient prognostic characteristics.19 Estimates of the life table relative hazard rate (RHR) were calculated using either the observed/expected (O/E) method for log-rank analyses or the exponentiated regression coefficient for Cox regression analyses.20
RESULTS
Immunophenotypic features of primary leukemic cells from children with CD2+CD19+ biphenotypic ALL.Within the final study population of 2,055 patients, there were 77 children (3.7%) with CD2+CD19+ biphenotypic ALL. The majority of the patients (N = 1631; 79.4%) were classified as BL, whereas 16.9% (N = 347) were classified as TL. Surface antigen profiles of leukemic cells from CD2+CD19+ biphenotypic patients were compared to those of leukemic cells in the BL and TL groups (Table 1). Leukemic cells from patients in the CD2+CD19+ and BL groups differed immunophenotypically for only a few antigens. Like the BL patients, CD2+CD19+ patients showed high expression frequency of CD10, CD19, CD34, and CD40 antigens; however, CD10 expression frequency was significantly lower (P < .0001) and CD34 expression was significantly higher (P = .0006) for the CD2+CD19+ group compared with the BL group. Neither group showed high expression frequencies for CD5 and CD7, although the CD5 comparison reached statistical significance (P = .04). Similarly, neither group exhibited high expression frequency for CD13 or CD33 myeloid antigens. Finally, as expected from the algorithm, higher expression frequency for CD2 occurred in the CD2+CD19+ group.
Immunophenotypic Features of Leukemic Cells From CD2+CD19+ Biphenotypic ALL Patients
| Variable . | CD2+CD19+ Biphenotypic ALL . | B-Lineage ALL . | P Value† . | T-Lineage ALL . | P Value‡ . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | N . | Mean* . | SE . | Med. . | N . | Mean . | SE . | Med. . | . | N . | Mean . | SE . | Med. . | . |
| CD2 | 77 | 58.9 | 2.7 | 56 | 1,631 | 10.4 | 0.2 | 8 | <.0001 | 347 | 75.0 | 1.6 | 88 | <.0001 |
| CD5 | 72 | 14.6 | 2.0 | 10 | 1,475 | 9.0 | 0.2 | 7 | .04 | 320 | 79.4 | 1.5 | 91 | <.0001 |
| CD7 | 64 | 16.3 | 2.7 | 7 | 1,427 | 8.8 | 0.2 | 7 | .24 | 316 | 86.5 | 0.8 | 92 | <.0001 |
| CD10 | 76 | 64.8 | 3.3 | 76 | 1,601 | 76.3 | 0.6 | 86 | <.0001 | 342 | 23.1 | 1.7 | 6 | <.0001 |
| CD19 | 77 | 83.5 | 1.7 | 88 | 1,631 | 85.0 | 0.3 | 89 | .74 | 347 | 5.2 | 0.4 | 2 | <.0001 |
| CD34 | 72 | 68.7 | 3.5 | 79 | 1,449 | 55.2 | 0.9 | 66 | .0006 | 319 | 36.2 | 2.0 | 23 | <.0001 |
| CD40 | 59 | 45.4 | 4.3 | 40 | 1,250 | 42.3 | 0.9 | 39 | .49 | 275 | 5.1 | 0.6 | 2 | <.0001 |
| CD13 | 36 | 16.0 | 3.6 | 6 | 758 | 13.0 | 0.7 | 4 | .72 | 235 | 13.3 | 1.6 | 3 | .11 |
| CD33 | 58 | 11.0 | 2.4 | 3 | 920 | 7.8 | 0.5 | 2 | .87 | 263 | 7.1 | 1.0 | 1 | .03 |
| Variable . | CD2+CD19+ Biphenotypic ALL . | B-Lineage ALL . | P Value† . | T-Lineage ALL . | P Value‡ . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | N . | Mean* . | SE . | Med. . | N . | Mean . | SE . | Med. . | . | N . | Mean . | SE . | Med. . | . |
| CD2 | 77 | 58.9 | 2.7 | 56 | 1,631 | 10.4 | 0.2 | 8 | <.0001 | 347 | 75.0 | 1.6 | 88 | <.0001 |
| CD5 | 72 | 14.6 | 2.0 | 10 | 1,475 | 9.0 | 0.2 | 7 | .04 | 320 | 79.4 | 1.5 | 91 | <.0001 |
| CD7 | 64 | 16.3 | 2.7 | 7 | 1,427 | 8.8 | 0.2 | 7 | .24 | 316 | 86.5 | 0.8 | 92 | <.0001 |
| CD10 | 76 | 64.8 | 3.3 | 76 | 1,601 | 76.3 | 0.6 | 86 | <.0001 | 342 | 23.1 | 1.7 | 6 | <.0001 |
| CD19 | 77 | 83.5 | 1.7 | 88 | 1,631 | 85.0 | 0.3 | 89 | .74 | 347 | 5.2 | 0.4 | 2 | <.0001 |
| CD34 | 72 | 68.7 | 3.5 | 79 | 1,449 | 55.2 | 0.9 | 66 | .0006 | 319 | 36.2 | 2.0 | 23 | <.0001 |
| CD40 | 59 | 45.4 | 4.3 | 40 | 1,250 | 42.3 | 0.9 | 39 | .49 | 275 | 5.1 | 0.6 | 2 | <.0001 |
| CD13 | 36 | 16.0 | 3.6 | 6 | 758 | 13.0 | 0.7 | 4 | .72 | 235 | 13.3 | 1.6 | 3 | .11 |
| CD33 | 58 | 11.0 | 2.4 | 3 | 920 | 7.8 | 0.5 | 2 | .87 | 263 | 7.1 | 1.0 | 1 | .03 |
Values are mean ± SE and median percentages of cells expressing the indicated antigen.
Kruskal-Wallis nonparametric rank test comparing CD2+CD19+ biphenotypic ALL and B-lineage ALL.
Kruskal-Wallis nonparametric rank test comparing CD2+CD19+ biphenotypic ALL and T-lineage ALL.
In contrast, leukemic cells of CD2+CD19+ patients differed immunophenotypically from leukemic cells of TL patients in almost every antigen category (Table 1). Median expression frequencies for the T-lineage differentiation antigens CD2, CD5, and CD7 were substantially lower (P < .0001 for each comparison) in biphenotypic patients compared with TL patients. In contrast, CD10, CD34, and CD40 expression frequencies were substantially higher (P < .0001 for each comparison) in biphenotypic patients compared with TL patients. As expected from the algorithm, higher expression frequency for CD19 occurred in CD2+CD19+ compared with TL patients.
Presenting features of children with CD2+CD19+ biphenotypic ALL.As was observed for the surface antigen profiles, presenting features of CD2+CD19+ patients generally were similar to BL patients, but showed numerous significant differences compared to TL patients (Table 2). Compared with BL patients, the CD2+CD19+ group had a significantly different age distribution (P = .01) primarily due to a higher percentage of children greater than 10 years of age (32.5% v 19.3%). WBC count was also significantly different (P = .04) with proportionately more CD2+CD19+ patients presenting with WBC counts in the lowest (<20,000/μL) range (74.0% v 60.5%) and fewer CD2+CD19+ patients presenting in the highest (≥50,000/μL) range (10.4% v 21.2%). Also, CD2+CD19+ patients more often had a mediastinal mass than BL patients (P = .02). Platelet count at diagnosis showed a highly significant difference (P = .002) with levels of CD2+CD19+ patients less frequently in the low range (<50,000/μL) and more frequently in the high range (≥150,000/μL). Within the subset of 35 CD2+CD19+ and 611 BL patients who had centrally reviewed cytogenetic analyses, no differences in karyotypic features were found.
Presenting Features of Children With CD2+CD19+ Biphenotypic ALL
| Variable . | Category . | CD2+CD19+ ALL (N = 77) . | B-Lineage ALL . | P Value* . | T-Lineage ALL . | P Value† . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | . | N . | (%) . | (N = 1,631) . | . | (N = 347) . | . | ||
| . | . | . | . | N . | (%) . | . | N . | (%) . | . |
| Age (yr) | <1 | 1 | (1.3) | 71 | (4.4) | .01 | 3 | (0.9) | .87 |
| 1-9 | 51 | (66.2) | 1,245 | (76.3) | 223 | (64.3) | |||
| >10 | 25 | (32.5) | 315 | (19.3) | 121 | (34.9) | |||
| WBC (×109/L) | 1-19 | 57 | (74.0) | 986 | (60.5) | .04 | 119 | (34.3) | <.0001 |
| 20-49 | 12 | (15.6) | 299 | (18.3) | 40 | (11.5) | |||
| ≥50 | 8 | (10.4) | 346 | (21.2) | 188 | (54.2) | |||
| Sex | Male | 39 | (50.6) | 860 | (52.7) | .81 | 248 | (71.5) | .0007 |
| Female | 38 | (49.4) | 771 | (47.3) | 99 | (28.5) | |||
| Race | White | 57 | (74.0) | 1,246 | (76.4) | .86 | 253 | (73.1) | .24 |
| Black | 3 | (3.9) | 66 | (4.0) | 32 | (9.2) | |||
| Other | 17 | (22.1) | 319 | (19.6) | 61 | (17.6) | |||
| Down syndrome | Yes | 1 | (1.3) | 37 | (2.3) | .87 | 5 | (1.4) | .66 |
| No | 76 | (98.7) | 1,592 | (97.7) | 341 | (98.6) | |||
| Liver | Normal | 45 | (58.4) | 770 | (47.3) | .13 | 152 | (44.3) | .03 |
| Mod. enlarg.‡ | 31 | (40.3) | 805 | (49.4) | 168 | (49.0) | |||
| Markedly enlarg. | 1 | (1.3) | 54 | (3.3) | 23 | (6.7) | |||
| Spleen | Normal | 42 | (54.5) | 742 | (45.5) | .30 | 138 | (39.8) | .01 |
| Mod. enlarg. | 32 | (41.6) | 807 | (49.5) | 161 | (46.4) | |||
| Markedly enlarg. | 3 | (3.9) | 82 | (5.0) | 48 | (13.8) | |||
| Lymph nodes | Normal | 46 | (59.7) | 923 | (56.6) | .22 | 108 | (31.1) | <.0001 |
| Mod. enlarg. | 31 | (40.3) | 647 | (39.7) | 149 | (42.9) | |||
| Markedly enlarg. | 0 | (0.0) | 61 | (3.7) | 90 | (25.9) | |||
| Mediastinal mass | Absent | 73 | (94.8) | 1,593 | (97.7) | .02 | 202 | (58.2) | <.0001 |
| Small | 2 | (2.6) | 32 | (2.0) | 40 | (11.5) | |||
| Large | 2 | (2.6) | 6 | (0.4) | 105 | (30.3) | |||
| Hemoglobin (g/dL) | 1-7.9 | 43 | (55.8) | 961 | (59.5) | .26 | 108 | (31.7) | <.0001 |
| 8.0.-10.9 | 30 | (39.0) | 508 | (31.5) | 120 | (35.2) | |||
| ≥11.0 | 4 | (5.2) | 146 | (9.0) | 113 | (33.1) | |||
| Platelets (×109/L) | 1-49 | 26 | (33.8) | 841 | (51.6) | .002 | 123 | (35.5) | .81 |
| 50-149 | 27 | (35.1) | 503 | (30.9) | 128 | (37.0) | |||
| ≥150 | 24 | (31.2) | 285 | (17.5) | 95 | (27.5) | |||
| CNS disease at diagnosis | Yes | 2 | (2.6) | 32 | (2.0) | .97 | 24 | (7.0) | .23 |
| No | 75 | (97.4) | 1,587 | (98.0) | 318 | (93.0) | |||
| NCI risk category | Standard | 44 | (57.1) | 1,007 | (61.7) | .49 | 105 | (30.3) | <.0001 |
| Poor | 33 | (42.9) | 624 | (38.3) | 242 | (69.7) | |||
| Karyotypic features | |||||||||
| 1.Number | Normal (46) | 13 | (37.1) | 159 | (26.0) | .45 | 60 | (39.5) | <.0001 |
| Hypodiploid (<46) | 3 | (8.6) | 38 | (6.2) | 4 | (2.6) | |||
| Psuedodiploid (46) | 5 | (14.3) | 159 | (26.0) | 70 | (46.1) | |||
| Hyperdiploid (47-50) | 4 | (11.4) | 66 | (10.8) | 16 | (10.5) | |||
| Hyperdiploid (>50) | 10 | (28.6) | 189 | (30.9) | 2 | (1.3) | |||
| 2.Aberrations | Normal | 13 | (37.1) | 159 | (26.0) | .21 | 60 | (39.5) | .95 |
| Abnormal | 22 | (62.9) | 452 | (74.0) | 92 | (60.5) | |||
| 3.Translocations | t(4; 11) present | 0 | (0.0) | 21 | (3.4) | .53 | 0 | (0.0) | >.99 |
| t(4; 11) absent | 35 | (100.0) | 590 | (96.6) | 152 | (100.0) | |||
| t(9; 22) present | 0 | (0.0) | 18 | (2.9) | .62 | 1 | (0.7) | .42 | |
| t(9; 22) absent | 35 | (100.0) | 593 | (97.1) | 151 | (99.3) | |||
| Variable . | Category . | CD2+CD19+ ALL (N = 77) . | B-Lineage ALL . | P Value* . | T-Lineage ALL . | P Value† . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | . | N . | (%) . | (N = 1,631) . | . | (N = 347) . | . | ||
| . | . | . | . | N . | (%) . | . | N . | (%) . | . |
| Age (yr) | <1 | 1 | (1.3) | 71 | (4.4) | .01 | 3 | (0.9) | .87 |
| 1-9 | 51 | (66.2) | 1,245 | (76.3) | 223 | (64.3) | |||
| >10 | 25 | (32.5) | 315 | (19.3) | 121 | (34.9) | |||
| WBC (×109/L) | 1-19 | 57 | (74.0) | 986 | (60.5) | .04 | 119 | (34.3) | <.0001 |
| 20-49 | 12 | (15.6) | 299 | (18.3) | 40 | (11.5) | |||
| ≥50 | 8 | (10.4) | 346 | (21.2) | 188 | (54.2) | |||
| Sex | Male | 39 | (50.6) | 860 | (52.7) | .81 | 248 | (71.5) | .0007 |
| Female | 38 | (49.4) | 771 | (47.3) | 99 | (28.5) | |||
| Race | White | 57 | (74.0) | 1,246 | (76.4) | .86 | 253 | (73.1) | .24 |
| Black | 3 | (3.9) | 66 | (4.0) | 32 | (9.2) | |||
| Other | 17 | (22.1) | 319 | (19.6) | 61 | (17.6) | |||
| Down syndrome | Yes | 1 | (1.3) | 37 | (2.3) | .87 | 5 | (1.4) | .66 |
| No | 76 | (98.7) | 1,592 | (97.7) | 341 | (98.6) | |||
| Liver | Normal | 45 | (58.4) | 770 | (47.3) | .13 | 152 | (44.3) | .03 |
| Mod. enlarg.‡ | 31 | (40.3) | 805 | (49.4) | 168 | (49.0) | |||
| Markedly enlarg. | 1 | (1.3) | 54 | (3.3) | 23 | (6.7) | |||
| Spleen | Normal | 42 | (54.5) | 742 | (45.5) | .30 | 138 | (39.8) | .01 |
| Mod. enlarg. | 32 | (41.6) | 807 | (49.5) | 161 | (46.4) | |||
| Markedly enlarg. | 3 | (3.9) | 82 | (5.0) | 48 | (13.8) | |||
| Lymph nodes | Normal | 46 | (59.7) | 923 | (56.6) | .22 | 108 | (31.1) | <.0001 |
| Mod. enlarg. | 31 | (40.3) | 647 | (39.7) | 149 | (42.9) | |||
| Markedly enlarg. | 0 | (0.0) | 61 | (3.7) | 90 | (25.9) | |||
| Mediastinal mass | Absent | 73 | (94.8) | 1,593 | (97.7) | .02 | 202 | (58.2) | <.0001 |
| Small | 2 | (2.6) | 32 | (2.0) | 40 | (11.5) | |||
| Large | 2 | (2.6) | 6 | (0.4) | 105 | (30.3) | |||
| Hemoglobin (g/dL) | 1-7.9 | 43 | (55.8) | 961 | (59.5) | .26 | 108 | (31.7) | <.0001 |
| 8.0.-10.9 | 30 | (39.0) | 508 | (31.5) | 120 | (35.2) | |||
| ≥11.0 | 4 | (5.2) | 146 | (9.0) | 113 | (33.1) | |||
| Platelets (×109/L) | 1-49 | 26 | (33.8) | 841 | (51.6) | .002 | 123 | (35.5) | .81 |
| 50-149 | 27 | (35.1) | 503 | (30.9) | 128 | (37.0) | |||
| ≥150 | 24 | (31.2) | 285 | (17.5) | 95 | (27.5) | |||
| CNS disease at diagnosis | Yes | 2 | (2.6) | 32 | (2.0) | .97 | 24 | (7.0) | .23 |
| No | 75 | (97.4) | 1,587 | (98.0) | 318 | (93.0) | |||
| NCI risk category | Standard | 44 | (57.1) | 1,007 | (61.7) | .49 | 105 | (30.3) | <.0001 |
| Poor | 33 | (42.9) | 624 | (38.3) | 242 | (69.7) | |||
| Karyotypic features | |||||||||
| 1.Number | Normal (46) | 13 | (37.1) | 159 | (26.0) | .45 | 60 | (39.5) | <.0001 |
| Hypodiploid (<46) | 3 | (8.6) | 38 | (6.2) | 4 | (2.6) | |||
| Psuedodiploid (46) | 5 | (14.3) | 159 | (26.0) | 70 | (46.1) | |||
| Hyperdiploid (47-50) | 4 | (11.4) | 66 | (10.8) | 16 | (10.5) | |||
| Hyperdiploid (>50) | 10 | (28.6) | 189 | (30.9) | 2 | (1.3) | |||
| 2.Aberrations | Normal | 13 | (37.1) | 159 | (26.0) | .21 | 60 | (39.5) | .95 |
| Abnormal | 22 | (62.9) | 452 | (74.0) | 92 | (60.5) | |||
| 3.Translocations | t(4; 11) present | 0 | (0.0) | 21 | (3.4) | .53 | 0 | (0.0) | >.99 |
| t(4; 11) absent | 35 | (100.0) | 590 | (96.6) | 152 | (100.0) | |||
| t(9; 22) present | 0 | (0.0) | 18 | (2.9) | .62 | 1 | (0.7) | .42 | |
| t(9; 22) absent | 35 | (100.0) | 593 | (97.1) | 151 | (99.3) | |||
Global chi-square test for homogeneity, B-lineage ALL v CD2+CD19+ biphenotypic ALL.
Global chi-square test for homogeneity, T-lineage ALL v CD2+CD19+ biphenotypic ALL.
Degree of organomegaly and size of mediastinal mass were determined as described in Materials and Methods.
In comparing presenting features of CD2+CD19+ to TL patients, a number of differences were observed (Table 2). WBC distribution showed a highly significant difference, because of the higher percentage of CD2+CD19+ patients in the lowest WBC range (74.0% v 34.3%). Also, CD2+CD19+ patients were less likely to be male than patients in the TL group (50.6% v 71.5%, P = .0006). Nonenlarged liver, spleen, and lymph nodes, and absence of a mediastinal mass were observed in 58.4%, 54.5%, 59.7%, and 94.8%, respectively, of the CD2+CD19+ patients as compared with 44.3%, 39.8%, 31.1%, and 58.2%, respectively, of the TL patients. All of these comparisons showed significant differences (P values were .03, .01, <.0001, and <.0001, respectively). Hemoglobin levels also differed significantly (P < .0001) for the CD2+CD19+ compared with the TL group: CD2+CD19+ patients were more often in the low (<8 g/dL) range (55.8% v 31.7%) and less often in the high (≥11 g/dL) range (5.2% v 33.1%). Because of the very large difference in WBC distribution for the CD2+CD19+ and TL patients, these groups also were significantly different in NCI risk classification (P < .0001), with 57.1% of CD2+CD19+ compared with 30.3% of TL patients assigned to the standard risk category. Cytogenetic comparisons of the groups showed a highly significant difference in chromosome number (P < .0001) with the CD2+CD19+ patients having a much greater proportion of high hyperdiploid (>50) presentations (28.6% v 1.3%), and a correspondingly lower proportion with pseudodiploid karyotypes (14.3% v 46.1%). There also were more CD2+CD19+ patients with hypodiploid presentations (8.6% v 2.6%), but this difference was based on very few patients.
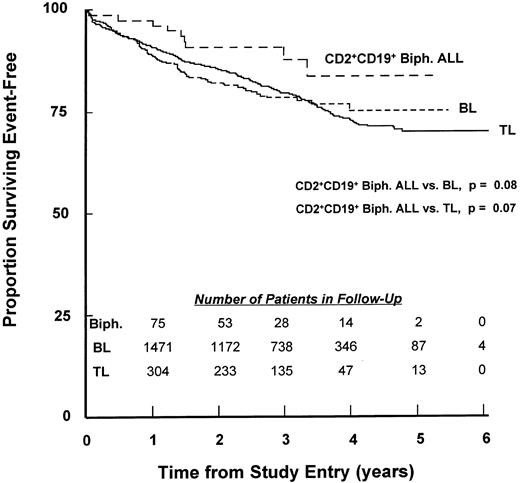
Treatment outcome of children with CD2+CD19+ biphenotypic ALL.Induction therapy results were similar for CD2+CD19+ patients compared to each of the other two groups, with 98.7%, 97.8%, and 97.3% of the CD2+CD19+, BL, and TL patients, respectively, achieving remission. However, results of a log-rank test of EFS outcome were suggestive (borderline significance) of an improved outcome for the CD2+CD19+ patients compared with either the BL or TL groups (CD2+CD19+v BL, P = .08; CD2+CD19+v TL, P = .07) (Fig 1). The 4-year EFS estimates were 83.7% (SD = 5.7%) for CD2+CD19+, 72.8% (SD = 1.4%) for BL, and 75.2% (SD = 3.0%) for TL. RHR estimates for BL and TL compared with CD2+CD19+ were 1.79 and 1.90, respectively. Because the CD2+CD19+ group had a significantly different presentation of several prognostic features compared with the other two groups, the independent influence of the CD2+CD19+ immunophenotype was assessed by a multivariate analysis using Cox regression. This analysis established age, WBC count, Down syndrome, central nervous system (CNS) disease at diagnosis, hemoglobin level, CD10 expression frequency, and lymph node enlargement as features that retained prognostic influence after adjustment. Immunophenotype (CD2+CD19+, BL, and TL) also was significant (likelihood ratio test, P = .001), but this was primarily due to the adjusted difference in outcome between the BL and TL groups, as recently reported.21 The multivariate adjusted RHR for BL compared to CD2+CD19+ patients was 1.43 (P = .29), which is reduced and no longer significant compared with the unadjusted estimate. The RHR for TL compared to CD2+CD19+ was 1.16 (P = .76), suggesting a major reduction in risk for TL patients once adjustment was made for the more favorable characteristics of the CD2+CD19+ group.
EFS of children with CD2+CD19+ biphenotypic ALL. Percentages of 77 biphenotypic (long hatches), 1,631 BL (short hatches), and 347 TL (solid line) patients achieving EFS during 6 years of follow-up were calculated as described in Materials and Methods. The number of patients in each group remaining in follow-up at the indicated time points is shown in the inset. P values for comparisons of the biphenotypic group to the BL and TL groups are based on the pattern of outcome across the entire period of patient follow-up.
EFS of children with CD2+CD19+ biphenotypic ALL. Percentages of 77 biphenotypic (long hatches), 1,631 BL (short hatches), and 347 TL (solid line) patients achieving EFS during 6 years of follow-up were calculated as described in Materials and Methods. The number of patients in each group remaining in follow-up at the indicated time points is shown in the inset. P values for comparisons of the biphenotypic group to the BL and TL groups are based on the pattern of outcome across the entire period of patient follow-up.
Additional analyses were performed to examine potentially interesting subsets of the CD2+CD19+ group. Leukemic cells from most of the CD2+CD19+ patients were assayed for the T-cell markers CD5 and/or CD7 (see Table 1). Within this group 19.4% showed positivity (≥30% of their cells expressed the indicated antigen) for CD5 and/or CD7. The EFS outcome in this CD2+CD19+ subgroup was not significantly different from the remaining CD2+CD19+ patients who were not CD5+ or CD7+ (P = .56). Leukemic cells from a subset of CD2+CD19+ patients also were assayed for the myeloid antigens CD13 and/or CD33 (see Table 1). A total of 13 patients were positive for one or both of these antigens. No difference in EFS outcome was noted for the CD2+CD19+ patients with myeloid antigen positivity compared to others within the CD2+CD19+ group (P = .89).
DISCUSSION
We have examined the clinical importance of CD2/CD19 coexpression by leukemic cells from a large cohort of newly diagnosed ALL patients enrolled in risk-adjusted treatment protocols of the CCG. In general, the prognostic features of CD2+CD19+ biphenotypic ALL patients were similar to those of BL patients, except that CD2+CD19+ patients more often presented with low WBC count and age ≥10 years, and more often had a mediastinal mass. In contrast, compared with the TL group, CD2+CD19+ patients generally presented with more favorable prognostic features including lower WBC level, absence of organomegaly, lack of mediastinal mass, lower hemoglobin level, and normal karyotype. The difference between groups was highly significant for each of these variables.
Patients in the CD2+CD19+ group also were similar to BL patients for expression of most of the cell surface antigens assayed. Indeed, both groups showed high expression frequencies for the B-lineage–associated antigens and showed low expression frequencies for the T-lineage (other than CD2) and myeloid antigens. However, it was notable that expression frequencies of CD10 and CD34 were lower and higher, respectively, for the CD2+CD19+ patients. In contrast, expression frequencies of almost all the antigens assayed were significantly different in CD2+CD19+ compared with TL patients. Expression frequencies of the other T-lineage–associated antigens CD5 and CD7 were lower for CD2+CD19+ compared with TL patients, and expression frequencies of the other B-lineage–associated antigens CD10, CD34, and CD40 were higher for CD2+CD19+ compared with TL patients. Taken together, these data suggest that CD2+CD19+ biphenotypic leukemic cells are more similar to B-lineage than T-lineage leukemic cells. These results are in accordance with our previously published observation that leukemic cells from biphenotypic CD2+CD19+ ALL patients are able to undergo in vitro differentiation to the pre-B stage, indicating that CD2/CD19 coexpression may be developmentally programmed and reflect a distinct stage of B-lymphocyte ontogeny.5 This notion is further supported by the existence in human fetal liver and BM of a small population of CD2+CD19+ biphenotypic lymphocyte precursors that are thought to represent the putative normal counterparts of CD2+CD19+ ALL cells.5
A limited number of studies have examined ALL patients whose leukemic cells exhibited coexpression of B- and T-lineage differentiation antigens.5-7,22,23 As mentioned above, during detailed immunophenotypic analyses of leukemic cells from 336 ALL patients, we had previously identified 5 CD2+CD19+ biphenotypic ALL patients.5 Four of these patients expressed several B-lineage differentiation antigens whereas 1 expressed several T-lineage differentiation antigens. All 5 patients achieved remission and 4 of 5 became long-term survivors. Hamdy et al22 identified two childhood ALL patients who expressed CD19 and/or CD10 along with a number of T-lineage–associated antigens. Philip et al23 identified three patients with biphenotypic B- and T-lineage ALL, but none of these patients coexpressed CD2 and CD19. Lim et al6 identified three patients (ages 8, 13, and 21 years) with CD2+CD19+ ALL. These patients, like those described in the current study, also expressed other B-lineage, but not T-lineage antigens. However, because of the small study size, few conclusions could be drawn concerning the significance of the immunophenotype. Subsequently, Lenormand et al7 identified the CD2+CD19+ immunophenotype in 16 cases (11 pediatric and 5 adult) of ALL. Presenting features of the pediatric patients generally were similar to those reported herein. Age of CD2+CD19+ children was similar to that of TL patients, but was significantly different compared to children with BL. Numerous other features (male:female ratio, WBC and hemoglobin levels, presence of mediastinal mass) were similarly favorable for biphenotypic ALL and BL, but significantly different and unfavorable for TL compared with the biphenotypic group. Regardless of CD2 expression, the 16 cases could be classified as BL according to nomenclature of France's Immunological Study Group for Leukemia (GEIL). Most cases (9 of 16) showed rearranged immunoglobulin genes, but only 1 case had a rearranged T-cell receptor (TCR) gene. The investigators concluded that the CD2+CD19+ patients represented a subgroup within BL. Complete remission was achieved for all but 2 adult patients. No comparisons of outcome were given, but 2- and 3-year actuarial disease-free survival for the biphenotypic patients (children + adults) were 88% ± 20% and 67% ± 41%, respectively.
The current study extends previous accounts of biphenotypic ALL by examining the incidence and clinical significance of CD2+CD19+ coexpression in a large cohort of patients. We show that patients with the CD2+CD19+ immunophenotype have an outcome that is not significantly different from either BL or TL patients after adjustment for presenting features by multivariate analyses. Thus, among children with ALL treated on contemporary CCG risk-adjusted protocols, those with the CD2+CD19+ immunophenotype generally present with favorable prognostic features and usually achieve good outcomes after treatment. To our knowledge, this report provides the first comprehensive characterization of CD2+CD19+ biphenotypic ALL in children.
Supported in part by research grants including CCG Chairman's Grant No. CA-13539, CA-51425, CA-42633, CA-42111, CA-60437, CA-27137 from the National Cancer Institute, National Institutes of Health. F.M.U. is a Stohlman Scholar of the Leukemia Society of America.
Address reprint requests to Fatih M. Uckun, MD, PhD, University of Minnesota, Children's Cancer Group ALL Biology Reference Laboratory, Biotherapy Institute, University of Minnesota Academic Health Center, 2625 Patton Rd, Roseville, MN 55113.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal