Abstract
The human blood group Rh antigens are expressed by proteins encoded by a pair of highly homologous genes located at chromosome 1p34-36. One of the genes (RHCE ) encodes Rh CcEe antigens, while the other (RHD) the D antigen. Point mutations in the RHCE gene generate the C/c and E/e polymorphisms, while it has been shown that an RHD gene deletion can generate the D-negative phenotype. We have analyzed intron 4 of the RHCE and RHD genes and have defined the site of an RHD-specific deletion located in this intron. Using a multiplex RHD typing assay, which combines a reverse polymerase chain reaction (PCR) primer, which straddles this RHD-specific sequence, and a pair of primers located in exon 10 of the RHD gene, we have analyzed 357 different genomic DNA samples derived from individuals expressing D+, D−, weak D, and partial D phenotypes. Of these, we have noted a significant discordance with our multiplex PCR assay in the D− phenotypes dCcee and dccEe (which have been previously described) and weak D phenotypes. Our results suggest that in five serologically D− individuals we have identified an apparently intact RHD gene. Sequence analysis of transcripts obtained from one of these individuals (of phenotype dCCee) illustrates the presence of full-length RHD transcripts, which have a point mutation at nucleotide 121 (C → T), which generates an in-frame stop codon (Gln41Stop). Thus, we describe a different molecular basis for generating the D− phenotype to the complete RHD gene deletion described previously. We also show that there are discordances with serotype and the multiplex assay in weak D and partial D phenotypes, indicating that the underlying molecular basis can be heterogeneous. Existing Rh D PCR assays assume the complete absence of the RHD gene in D− phenotypes. We describe a different molecular basis for generating the D− phenotype to the complete RHD gene deletion described previously.
THE BLOOD GROUP Rh antigens are encoded by a pair of highly homologous genes (RHCE and RHD), which lie in tandem on chromosome 1p34-36.1-3 Their gene products, the Rh polypeptides are hydrophobic, unglycosylated, polytopic erythrocyte membrane proteins of which two full-length cDNA species have been characterized.4-8 One species of mRNA encodes Rh cE antigens,4,5,9,10 while the other encodes the D antigen.6,7,10 Rh Cc and Ee antigens have been predicted to be expressed on different polypeptide chains, based on immunochemical analysis,11-13 by a mechanism of alternate splicing (exon skipping).9,14 Rh Cc transcripts are proposed to lack exon 5, which encodes residues critical for Rh E/e antigenicity.9 Recent evidence refutes this hypothesis. A full-length Rh cE transcript was expressed in K562 cells, with resultant expression of both Rh c and E antigens.10 In addition, immunopurified Rh Cc proteins have been shown to be comprised of full-length, not truncated polypeptide chains.15
The genomic arrangement of the RHCE gene has been partially established, and is a 10 exon gene encompassing 75 Kb.16 The arrangement of the RHD gene is thought to be similar, and to be completely deleted in the majority of Caucasian D− chromosomes,1 but present or partially deleted in others.17,18 The extent of the deletion and precise molecular mechanisms underlying the D− phenotype in these individuals remains unknown. Several structural differences between the RHD and the RHCE gene are known. These differences are used in Rh D polymerase chain reaction (PCR) typing assays in the clinical management of hemolytic disease of the newborn (HDN) resulting from placental transfer of maternal anti-D. These assays are dependent on the complete absence of the region of the RHD gene being detected in D− individuals. Bennett et al19 have used the fact that the RHD gene has a larger exon 10 than the RHCE gene to type fetally-derived DNA. Others have used the fact that the RHCE gene has a larger intron 4 than the RHD gene,7,20 and also that sequence differences exist between exon 7 of the RHD and RHCE genes.21 Discrepancies between standard serological typing and PCR typing have been observed,20,22 23 with both false positives and false negatives being described.
In this report we describe a multiplex RHD typing assay based on amplification of RHD intron 4 and exon 10. We have validated this assay by screening 357 DNAs derived from individuals expressing different Rh phenotypes. We have identified that a significant proportion of individuals who express the D− haplotypes dCe and dcE, weak D and partial D phenotypes give discrepant results within the assay, indicating that these individuals most probably possess partially deleted or rearranged RHD genes. We have identified an individual expressing the Rh phenotype dCe who possesses an intact RHD gene with a nonsense mutation altering codon 41 (Gln → stop). Our findings show the importance of utilizing at least two different RHD typing assays in prenatal determination of fetal Rh D type, directed at different regions of the RHD gene.
MATERIALS AND METHODS
Sources of erythrocytes and extraction of genomic DNA (gDNA) from peripheral blood (PB) and fetal amniocytes.gDNA was extracted from 5 to 10 mL of PB and 5 mL amniotic fluid as described by Avent and Martin.24 The Rh DCcEe status of all erythrocytes was established by using routine D and CcEe typing antibodies: English samples: MAD-2 (BioProducts laboratory, Elstree, Hert); and Seraclone anti-D blend (BS 221, H41-11B7, BS 232; Biotest, Dreieich, Germany); Scottish samples: LDM1 plus blended anti-D ESD1 and LDM3 (Scottish National Blood Transfusion Service National reagents programme, Edinburgh, Scotland). Weak D phenotypes were established by demonstration of weakened D antigen expression by reaction profiles with MAD-2 and LDM1, and confirmed using the blended anti-Ds. D-variant erythrocytes were made available from either frozen cells or fresh samples referred to IBGRL, while others were made available through the third international workshop on monoclonal antibodies against red cell and related antigens (September 1996, Nantes, France) and were classified by typing using monoclonal anti-D into distinct D categories.
Cloning and DNA sequence analysis of RHD and RHCE intron 4 PCR products.A 1,200-bp PCR product containing intron 4 of the RHCE gene was amplified by using an exon 4 sense primer (5′-ACTTCTACGTGTTCGCAGCCTATTT-3′ ) and an exon 5 antisense primer (5′-TCTTCCTTTGGATTGGACTTCTCAG-3′ ) of the RHD and RHCE genes with gDNA derived from lymphocytes obtained from an individual of Rh phenotype dce. A 600-bp PCR product containing intron 4 of the RHD gene was obtained from lymphocytes derived from an individual of Rh phenotype DcE with the same primer pair as above. PCR conditions were as follows 94°C/1 min; 60°C/1 min, 30 s; 72°C/2 min, 30 s for 30 cycles. Fifty microliters of PCR reaction mix contained 2.5 mmol/L MgCl2 : 10 mmol/L Tris-Cl pH 8.3; 500 mmol/L KCl; 1 μmol/L each primer, 1.25 mmol/L each dNTP and 2.25 U Taq DNA polymerase (Perkin Elmer, Warrington, UK). Both PCR products were gel purified on Low-Melt point gels (Flowgen Instruments Ltd, Lichfield, UK), and cloned into pCRII (TA cloning kit, Invitrogen, San Diego, CA) using the manufacturers' instructions. DNA sequencing was performed using dye-labeled terminator cycle sequencing chemistry on an Applied Biosystems 373A DNA sequencer with 0.5 to 1.0 μg plasmid DNA as template. Both strands of the DNA were sequenced.
RHD gene intron 4/exon 10 multiplex assay.The multiplex PCR contains five primers, two specific for the RHD gene, one specific for the RHCE gene, and two primers common to both genes. A schematic representation of the PCR is shown in Fig 1. The primer sequences are as follows: RHD specific intron 4 (antisense) 5′-GAACCTGCTCTGTGAAGTGCT-3′ (Primer A) RHD specific exon 10 (antisense) 5′-CTCCTCAAAGAGTGGCAGAGAAAG-3′ (Primer B) RHCE specific intron 4 (antisense) 5′-GCTCACTGCCCGATTTTCTAT-3′ (Primer C) RHD/RHCE exon 10 (sense) 5′-CCTCATTTGGCTGTTGGATTTTA-3′ (Primer D) RHD/RHCE exon 4 (sense) 5′-CCTATTTTGGGCTGTACTGTGG-3′. (Primer E) PCR was performed in a reaction volume of 25 or 50 μL composed of the following: 2.5 mmol/L MgCl2 , 10 mmol/L Tris-Cl pH 8.3, 500 mmol/L KCl, 0.5 μmol/L primer A; 5 μmol/L primer B; 0.5 μmol/L primer C; 5 μmol/L primer D; 0.5 μmol/L primer E; 1.25 mmol/L each dNTP and 2.5 U Amplitaq DNA polymerase (Perkin Elmer). Cycling was 94°C/1 min; 68°C/1 min; 72°C/1 min, 30 s for 30 cycles on either a Perkin Elmer TC1 or 2400 thermal cycler. Following the completion of the PCR, the 50- or 25-μL reaction volume was reduced to approximately 20 μL by vacuum desiccation (after removing from under mineral oil if present), and products separated on a 6% (wt/vol) polyacrylamide gel.
Schematic representation of RHD Intron 4/exon 10 multiplex PCR. A diagrammatic representation of the multiplex assay used in this study is depicted in this figure. The approximate positions of the amplimers are shown with the identification letters (A through D) shown at the 5′ end of the primer. The sequences of the primers are shown and their nucleotide positions (numbered within either RHCE or RHD intron 4 (Fig 2); or corresponding to Rh cE4 5 or D6 cDNAs). The location of the RHD specific deletion or RHCE specific insertion are highlighted on the figure.
Schematic representation of RHD Intron 4/exon 10 multiplex PCR. A diagrammatic representation of the multiplex assay used in this study is depicted in this figure. The approximate positions of the amplimers are shown with the identification letters (A through D) shown at the 5′ end of the primer. The sequences of the primers are shown and their nucleotide positions (numbered within either RHCE or RHD intron 4 (Fig 2); or corresponding to Rh cE4 5 or D6 cDNAs). The location of the RHD specific deletion or RHCE specific insertion are highlighted on the figure.
Reverse-transcription (RT) coupled with PCR amplification of Rh transcripts from PB.Full-length Rh transcripts were isolated following RT-PCR using total RNA isolated from whole blood by the ammonium-chloride/bicarbonate lysis method.25 Six micrograms of total RNA was used as template for first-strand cDNA synthesis using oligo d(T20) as primer, and AMV-reverse transcriptase as described by Sambrook et al.26 The resultant cDNA was resuspended in 50 μL of H2O, and 1 μL used for PCR with forward and reverse amplimers complementary to the 5′ and 3′ noncoding regions of the Rh mRNAs. The amplimer sequences were: forward primer (5′-TCCCTCAAGCCCTCAAGTAG-3′ ) and reverse primer (5′-GTACAAATGCAGGCAACAGTG-3′ ), corresponding to nts −81 to −56 and 1328-1308 respectively of the Rh30A cDNA sequence. PCR amplification was performed using the following conditions: 94°C for 1 minute, 55°C for 1 minute, and 72°C for 3 minutes for 35 cycles in a 50-μL reaction mix composed of the following: 10 mmol/L Tris-HCl pH 8.3, 500 mmol/L KCl, 2.5 mmol/L MgCl2 1 μmol/L each primer 1.25 mmol/L each dNTP and 1.25 U Bio-X-Act DNA polymerase cocktail (Bioline, London, UK). PCR products corresponding to full-length Rh transcripts were gel-purified on low-melt point agarose gels and cloned into pCRII vector using the manufacturer's instructions. Clones were confirmed as having inserts by PCR, and were classified as either RHCE or RHD transcripts by HincII/EcoRI double digestion. Two Rh D and two Rh Ce clones were sequenced fully on both strands using an Applied-Biosystems 373A DNA sequencer.
RESULTS
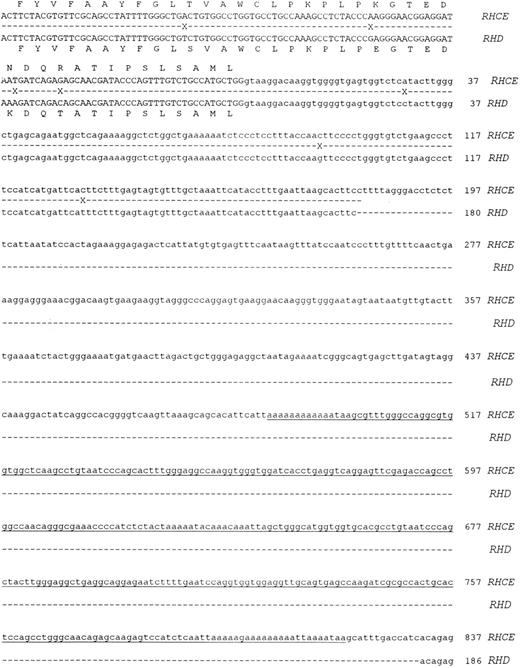
Molecular cloning of intron 4 of the RHCE and RHD genes.Genomic clones corresponding to intron 4 of the RHCE and RHD genes were isolated as described in Materials and Methods. One RHCE and three RHD clones were isolated and fully sequenced on both strands. The DNA sequences obtained were identical for all RHD clones, and the RHD intron 4 was found to be composed of 426 nucleotides. The RHCE intron 4 was composed of 1077 nucleotides, and contains an Alu repeat element. The extent of the Alu repeat element (including both poly A rich tracts located at ends of the head to tail dimeric repeats) is underlined in Fig 2. The aligned sequences of both RHD and RHCE intron 4 are illustrated in Fig 2. Analysis of the DNA sequence indicated that the exon 4/intron 4 and intron 4/exon 5 boundaries differed to those described by Cherif-Zahar et al,16 (Fig 3) in all clones analyzed (five). DNA sequence electropherograms obtained on the ABI-373A DNA sequencer for the intron/exon boundaries are displayed in Fig 3.
Nucleotide sequences of cloned RHCE and RHD gene Intron 4. The complete nucleotide sequences of the RHCE and RHD gene intron 4 are displayed. The sequences are derived from cloned PCR products, the sequences of exon 4 and 5 are displayed in uppercase. Intron 4 sequence is displayed as lowercase, and the first nucleotide of the intron is numbered as position 1. The RHCE intron extends to 1077 nts, while the RHD intron to 426 nts. Sequence differences between the two genes are denoted by a cross on the shadow sequence. The two nucleotide sequences have been submitted to Genbank/EMBL databases and have the following accession numbers: Intron 4 RHCE:Y10604, intron 4 RHD:Y10605.
Nucleotide sequences of cloned RHCE and RHD gene Intron 4. The complete nucleotide sequences of the RHCE and RHD gene intron 4 are displayed. The sequences are derived from cloned PCR products, the sequences of exon 4 and 5 are displayed in uppercase. Intron 4 sequence is displayed as lowercase, and the first nucleotide of the intron is numbered as position 1. The RHCE intron extends to 1077 nts, while the RHD intron to 426 nts. Sequence differences between the two genes are denoted by a cross on the shadow sequence. The two nucleotide sequences have been submitted to Genbank/EMBL databases and have the following accession numbers: Intron 4 RHCE:Y10604, intron 4 RHD:Y10605.
Nucleotide sequences of exon 4/Intron 4 and Intron 4/exon 5 boundaries. Nucleotide sequences obtained at the intron/exon boundaries of both cloned RHCE and RHD genes are displayed. Exonic sequences are shown in uppercase, intronic sequences in lowercase. The last nucleotide of exon 4 and the first of exon 5 are numbered. The numbering corresponds to the Rh cE and D cDNAs with nucleotide 1 being the first base of the ATG triplet. A representative sequence electropherogram generated during sequence analysis of these regions is displayed in this figure.
Nucleotide sequences of exon 4/Intron 4 and Intron 4/exon 5 boundaries. Nucleotide sequences obtained at the intron/exon boundaries of both cloned RHCE and RHD genes are displayed. Exonic sequences are shown in uppercase, intronic sequences in lowercase. The last nucleotide of exon 4 and the first of exon 5 are numbered. The numbering corresponds to the Rh cE and D cDNAs with nucleotide 1 being the first base of the ATG triplet. A representative sequence electropherogram generated during sequence analysis of these regions is displayed in this figure.
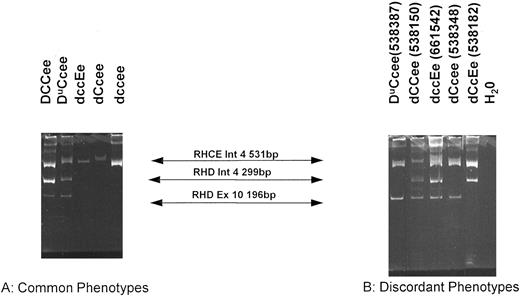
Organization of the RHD gene in individuals different Rh phenotypes as determined by the RHD exon 10/intron 4 multiplex assay.The multiplex assay was performed on a number of genomic DNA samples derived from lymphocytes and amniocytes isolated from individuals of known Rh phenotype. Three bands were obtained in D+ samples (531 bp; 299 bp and 196 bp) and only one band was obtained in D− samples (531 bp) (see Figs 1 and 4). These results are tabulated in Table 1. A typical PCR using the multiplex PCR is displayed in Fig 4. Significantly, the assay showed concordance with RhD serotype in all D+ samples tested, except in partial D and weak D phenotypes (Tables 1 and 2). However, the assay was discordant in some D− phenotypes (Tables 1 and 2). The most common D− phenotype (dce) was concordant with both the exon 10 and intron 4 bands, but D− haplotypes dCe and dcE showed marked discordance (Table 2). Of 33 dCcee samples, 8 were discordant with one or more aspect of the multiplex assay. Of these, 6 were exon 10 positive intron 4 negative, one was exon 10 negative intron 4 positive, while two were both exon 10 and intron 4 positive. Of the discordant dccEe phenotypes, two were both intron 4 and exon 10 positive (Table 2). There were five instances when both exon 10 and intron 4 bands were detected in these D− phenotypes. One of these donors (of dCCee phenotype, donor 538150) was further characterized after obtaining fresh samples of blood. Donor 538150 was reconfirmed as expressing the Rh phenotype dCCee, and a second genomic DNA sample gave the same discrepant result with the multiplex assay (Fig 4). Total RNA was extracted as described in Materials and Methods and Rh D transcripts isolated following RT-PCR (see below). Full-length Rh PCR products were cloned into pCRII and subjected to DNA sequence analysis. Two Rh D and three Rh Ce clones were sequenced fully. Both Rh D sequences were identical, as were all three Rh Ce transcripts. Rh D transcripts were found to possess a mutation at nucleotide 121 (C → T), which generates an in-frame stop codon (Gln41stop; Fig 5). Three other mutations were found in both Rh D transcripts, at codons 215 (TTC → CTC; Phe → Leu), 216 (TTG → CTG; Leu → Leu), and 330 (TAC → CAC; Tyr → His) (see Fig 4).
Multiplex PCR analysis of the RHD gene. This figure shows the products of the multiplex reaction after separation on 6% acrylamide gels. Products of 531 bp (RHCE intron 4); 299 bp (RHD intron 4) and 196 bp (RHD exon 10). Products are indicated between the two panels. (A) Illustrates samples that are concordant (all normal D+ and dce phenotypes; representing the majority of all samples tested) and (B) illustrates discordant samples. The Rh phenotype of each sample is indicated above each track. All samples on (A) and those on (B) were run on the same gel.
Multiplex PCR analysis of the RHD gene. This figure shows the products of the multiplex reaction after separation on 6% acrylamide gels. Products of 531 bp (RHCE intron 4); 299 bp (RHD intron 4) and 196 bp (RHD exon 10). Products are indicated between the two panels. (A) Illustrates samples that are concordant (all normal D+ and dce phenotypes; representing the majority of all samples tested) and (B) illustrates discordant samples. The Rh phenotype of each sample is indicated above each track. All samples on (A) and those on (B) were run on the same gel.
RHD Typing Using the Exon 10/Intron 4 Multiplex Assay (n = 357)
| . | . | Exon 10 . | Intron 4 . | ||
|---|---|---|---|---|---|
| . | . | + . | − . | + . | − . |
| D+ Phenotypes | DCCee (n = 39) | 39 | 0 | 39 | 0 |
| DccEE (n = 27) | 27 | 0 | 27 | 0 | |
| Dccee (n = 6) | 6 | 0 | 6 | 0 | |
| DCCEE (n = 1) | 1 | 0 | 1 | 0 | |
| DCcEe (n = 5) | 5 | 0 | 5 | 0 | |
| DCcee (n = 26) | 26 | 0 | 26 | 0 | |
| DccEe (n = 17) | 17 | 0 | 17 | 0 | |
| D− Phenotypes | dccee (n = 55) | 0 | 55 | 0 | 55 |
| dCcee (n = 33) | 7 | 26 | 3 | 30 | |
| dccEe (n = 43) | 2 | 41 | 2 | 41 | |
| dCCee (n = 4) | 1 | 3 | 1 | 3 | |
| dccEE (n = 3) | 0 | 3 | 0 | 3 | |
| dCcEe (n = 1) | 0 | 1 | 1 | 0 | |
| dccEe G+ (n = 1) | 1 | 0 | 0 | 1 | |
| Rhnull (n = 3) | 2 | 1 | 2 | 1 | |
| Du/D-Variant | DuCcee (n = 35) | 35 | 0 | 31 | 4 |
| phenotypes | DuccEe (n = 9) | 9 | 0 | 9 | 0 |
| DIICcee (n = 1) | 1 | 0 | 1 | 0 | |
| DIIICcee (n = 5) | 5 | 0 | 5 | 0 | |
| DIIIccee (n = 2) | 2 | 0 | 2 | 0 | |
| DIVaCcee (n = 1) | 1 | 0 | 1 | 0 | |
| DIVbCcee (n = 3) | 3 | 0 | 3 | 0 | |
| DVaCcee (n = 3) | 3 | 0 | 3 | 0 | |
| DVbCcee (n = 1) | 1 | 0 | 1 | 0 | |
| DVICcee (n = 9) | 9 | 0 | 0 | 9 | |
| DVIccEe (n = 5) | 5 | 0 | 0 | 5 | |
| DVIIccee (n = 7) | 7 | 0 | 7 | 0 | |
| DBT (n = 1) | 1 | 0 | 1 | 0 | |
| DFR (n = 2) | 2 | 0 | 1 | 1 | |
| DHMi (n = 4) | 4 | 0 | 4 | 0 | |
| DHMii (n = 2) | 2 | 0 | 2 | 0 | |
| DHR* (n = 1) | 1 | 0 | 1 | 0 | |
| DNU† (n = 1) | 1 | 0 | 1 | 0 | |
| DHAR (n = 1) | 0 | 1 | 0 | 1 | |
| . | . | Exon 10 . | Intron 4 . | ||
|---|---|---|---|---|---|
| . | . | + . | − . | + . | − . |
| D+ Phenotypes | DCCee (n = 39) | 39 | 0 | 39 | 0 |
| DccEE (n = 27) | 27 | 0 | 27 | 0 | |
| Dccee (n = 6) | 6 | 0 | 6 | 0 | |
| DCCEE (n = 1) | 1 | 0 | 1 | 0 | |
| DCcEe (n = 5) | 5 | 0 | 5 | 0 | |
| DCcee (n = 26) | 26 | 0 | 26 | 0 | |
| DccEe (n = 17) | 17 | 0 | 17 | 0 | |
| D− Phenotypes | dccee (n = 55) | 0 | 55 | 0 | 55 |
| dCcee (n = 33) | 7 | 26 | 3 | 30 | |
| dccEe (n = 43) | 2 | 41 | 2 | 41 | |
| dCCee (n = 4) | 1 | 3 | 1 | 3 | |
| dccEE (n = 3) | 0 | 3 | 0 | 3 | |
| dCcEe (n = 1) | 0 | 1 | 1 | 0 | |
| dccEe G+ (n = 1) | 1 | 0 | 0 | 1 | |
| Rhnull (n = 3) | 2 | 1 | 2 | 1 | |
| Du/D-Variant | DuCcee (n = 35) | 35 | 0 | 31 | 4 |
| phenotypes | DuccEe (n = 9) | 9 | 0 | 9 | 0 |
| DIICcee (n = 1) | 1 | 0 | 1 | 0 | |
| DIIICcee (n = 5) | 5 | 0 | 5 | 0 | |
| DIIIccee (n = 2) | 2 | 0 | 2 | 0 | |
| DIVaCcee (n = 1) | 1 | 0 | 1 | 0 | |
| DIVbCcee (n = 3) | 3 | 0 | 3 | 0 | |
| DVaCcee (n = 3) | 3 | 0 | 3 | 0 | |
| DVbCcee (n = 1) | 1 | 0 | 1 | 0 | |
| DVICcee (n = 9) | 9 | 0 | 0 | 9 | |
| DVIccEe (n = 5) | 5 | 0 | 0 | 5 | |
| DVIIccee (n = 7) | 7 | 0 | 7 | 0 | |
| DBT (n = 1) | 1 | 0 | 1 | 0 | |
| DFR (n = 2) | 2 | 0 | 1 | 1 | |
| DHMi (n = 4) | 4 | 0 | 4 | 0 | |
| DHMii (n = 2) | 2 | 0 | 2 | 0 | |
| DHR* (n = 1) | 1 | 0 | 1 | 0 | |
| DNU† (n = 1) | 1 | 0 | 1 | 0 | |
| DHAR (n = 1) | 0 | 1 | 0 | 1 | |
DHR: D variant due to point mutation in exon 5 of the RHD gene (J.W. Jones et al, submitted).
DNU: D variant due to point mutation in exon 7 of the RHD gene.27
Multiplex analysis of genomic DNA derived from 357 different individuals expressing different Rh phenotypes. This table illustrates the results of screening genomic DNAs with the multiplex assay with 357 genomic DNAs of which the Rh phenotypes of the erythrocytes had been previously established. This table also illustrates the phenotypes and numbers of samples that are positive or negative with the two RHD-specific aspects of the assay. Discordances in the assay from the serotype are highlighted in bold (further defined in Table 2).
Discordant Samples With the Multiplex RHD Assay
| Sample . | Phenotype . | Exon 10 . | Intron 4 . |
|---|---|---|---|
| 538184 | dCcee | + | + |
| 538348 | dCcee | + | + |
| 538381 | dCcee | + | − |
| 538385 | dCcee | + | − |
| 538388 | dCcee | + | − |
| 538401 | dCcee | + | − |
| 538437 | dCcee | + | − |
| 538182 | dCcee | − | + |
| 538150 | dCCee | + | + |
| 538364 | dccEe | + | + |
| 661542 | dccEe | + | + |
| 538374 | dCcEe | − | + |
| 538387 | DuCcEe | + | − |
| 538385 | DuCcEe | + | − |
| 538421 | DuCcEe | + | − |
| 538484 | DuCcEe | + | − |
| Sample . | Phenotype . | Exon 10 . | Intron 4 . |
|---|---|---|---|
| 538184 | dCcee | + | + |
| 538348 | dCcee | + | + |
| 538381 | dCcee | + | − |
| 538385 | dCcee | + | − |
| 538388 | dCcee | + | − |
| 538401 | dCcee | + | − |
| 538437 | dCcee | + | − |
| 538182 | dCcee | − | + |
| 538150 | dCCee | + | + |
| 538364 | dccEe | + | + |
| 661542 | dccEe | + | + |
| 538374 | dCcEe | − | + |
| 538387 | DuCcEe | + | − |
| 538385 | DuCcEe | + | − |
| 538421 | DuCcEe | + | − |
| 538484 | DuCcEe | + | − |
Discordant samples with the multiplex RHD assay. All genotypically discordant samples are illustrated. The donor identification number, serological phenotype and result of analysis with the two RHD-specific aspects of the assay are shown. Some discordant samples are also illustrated in Fig 4.
Nucleotide sequence of Rh D transcripts isolated from donor 538150. The nucleotide sequence obtained after DNA sequence analysis of 538150 RHD transcripts is shown. The sequence obtained from two different full-length cDNA clones was found to be identical.
Nucleotide sequence of Rh D transcripts isolated from donor 538150. The nucleotide sequence obtained after DNA sequence analysis of 538150 RHD transcripts is shown. The sequence obtained from two different full-length cDNA clones was found to be identical.
Significant discordance between serotype and multiplex assay were noted when gDNA derived from individuals expressing weak D and partial D phenotypes were used, which were obtained from both Aberdeen and Bristol blood transfusion centers, and from the third international workshop on monoclonal antibodies against red cell antigens. Four samples expressing the haplotype DuCe were shown to be RHD intron 4 negative, RHD exon 10 and RHCE intron 4 positive (Table 2; one example shown in Fig 4). Of these four samples, all were known not to be of DVI phenotype based on serological activity of these red cells with monoclonal antibodies. Thirteen samples of DVICe and DVIcE haplotypes were RHD intron 4 negative, as was 1 out of 2 samples expressing the DFR phenotype (Table 1), but retained the RHD exon 10 and RHCE intron 4 bands (not shown). One example of the RHaro phenotype was shown to be negative with both RHD aspects of the multiplex assay (intron 4 and exon 10) but with a positive RHCE intron 4 band (data not shown).
DISCUSSION
Significant progress has been made in the last 6 years in defining the molecular basis of human blood group antigen expression. Many human blood group antigens result from point mutations generating allelic forms of the genes encoding blood group active proteins.28 However, the Rh D positive/negative polymorphism is thought to be generated by the complete absence of the Rh D protein, initially shown to result from a complete RHD gene deletion.1 This finding shows that the absence of the Rh D protein is not physiologically detrimental to D− individuals. Therefore, it follows that RHD gene rearrangements (gene conversions, point mutations) which do not alter the Rh D protein structure significantly may be expressed at the erythrocyte surface. These RHD gene alterations generate partial D (D-variant) phenotypes, many of which have been defined at the molecular level.29 30 We have investigated the gross organization of the RHD gene in all commonly occurring Rh phenotypes including partial D and weak D (Du) phenotypes. Our findings suggest genetic diversity among dCe, dcE, and weak D phenotypes.
The molecular determination of blood group antigen expression has allowed the use of antenatal PCR-based detection of fetal blood groups in cases of maternal alloimmunization.19-21 We have optimized a multiplex RHD typing assay (analyzing intron 4 and exon 10 of the RHD gene), which we routinely use for prenatal Rh D determination, and have used this assay in this study. Our findings illustrate that a multiplex approach is essential for RHD typing; there are many situations where the exclusive use of a single RHD typing assay would generate both false negatives and positive results which confirms the findings of others.20,22 23
DNA sequences of RHD and RHCE intron 4.We report here the entire sequence of both the RHD and RHCE gene intron 4. The RHD gene intron is composed of 426 nucleotides, while the RHCE gene of 1077 nucleotides (Fig 2). The RHCE intron contains an Alu repeat element. These elements are widely dispersed in mammalian genomes.31,32 The absence of such a repeat from the RHD gene could potentially be why this gene serves as the acceptor of gene conversion events. Inhibition of gene conversion events by Alu repeat elements has been suggested for α1-α2 and β-γ globin genes,33,34 and this may explain why the RHCE gene serves as the donor in the majority of Rh gene conversion events generating partial D phenotypes involving RHD exons 4 and 5. One exception to this occurs in the RHaro phenotype, where an RHCE gene has an RHD-like exon 5.35 It is possible that a double crossing over event rather than gene conversion generates this phenotype.35 The sequence of a hybrid RHD gene intron 4 derived from an individual expressing the Rh phenotype DCW-has been recently determined.36 Analysis of Rh transcripts from this individual reveals that a hybrid RHCW-D gene comprising of exons 1 from a CW gene, exons 2-10 from an RHD gene. The RHD intron 4 sequence is identical to that described here except for two nucleotides, 320 and 339 (T → A and A → G, respectively).
We have determined the intron/exon boundaries of the exon 4/intron 4 and intron 4/exon 5 junctions of cloned PCR products from three different individuals (Fig 3). These sequences differ to those described by Cherif-Zahar et al.16 We have shown that exons 4 and 5 of both genes are composed of 150 and 195 nucleotides, respectively. We assume that the sequences derived from a single cosmid clone for the exon 4/intron 4 and intron 4/exon 5 boundaries reported by these workers are erroneous, possibly caused by band compression of these GC-rich sequences.36 The intron 4/exon 5 splice boundary described here agrees with that determined by Huang36 from 15 different individuals.
We have cloned and sequenced intron 4 of the RHCE and RHD genes primarily to identify the site of the RHD-specific deletion identified originally by Arce et al.7 Primers complementary to this RHD-specific site have been incorporated into a multiplex PCR-based assay for determination of RHD type. The intron 4 assay initially described by Arce et al,7 involved the use of amplimers which are common to both RHCE and RHD genes, and amplified two products (600 and 1,200 bp) from D+ genomes and one product (1,200 bp) from D−. As a consequence, in D+ genomes often the only product observed under certain reaction conditions (eg, low concentrations of DNA, frequently obtained from amniotic fluid samples) is the 600-bp RHD-specific band (N.D.A. and P.G.M.; S.S.A.F. and Susan Cochrane, independent unpublished observations, September 1995). We have shown that by using an entirely RHD-intron 4 specific primer that a more robust assay is obtained, and is controlled with the incorporation of a RHCE-specific amplimer. Exon 10 of the RHD gene is also amplified using an RHD-specific reverse amplimer (Fig 1).
RHD genes and gene fragments in D− genomes.Using the RHD intron 4/exon 10 multiplex assay and genomic DNA derived from individuals expressing D− phenotypes, we have found that the dce haplotype is completely concordant with the assay, supporting the hypothesis that a complete RHD gene deletion generates this phenotype.1 However, in the D− haplotypes dCe and dcE we have illustrated that a significant proportion of these individuals carry portions of the RHD gene, and probably an intact but dysfunctional RHD gene in five individuals (phenotypes 2 × dCcee, dCCee and 2 × dccEe; Table 2). Of the 33 individuals studied who express the products of the dCe gene complex, 7 out of 33 (21.2%) showed discordant results with the multiplex assay. In individuals who express the dcE gene complex, 2 out of 43 (4.6%) were discordant (Tables 1 and 2). Analysis of Rh transcripts from one of these individuals (538150, expressing the Rh phenotype dCCee) by RT-PCR showed that intact RHD transcripts were present, and that a point mutation in the RHD gene at codon 41 generating a stop codon is the molecular basis of the D− phenotype in this individual (Fig 6). This finding suggests that the mutant dCe RHD gene arose by point mutation of a DCe chromosome. The majority of dCe and dcE haplotype genomes appear to lack all portions of the RHD gene, and therefore probably have arisen by a similar RHD gene deletion event to that observed in dce chromosomes. Other workers have described the presence of RHD genes or gene fragments in D− individuals.17,18 In the dCes haplotype there is a hybrid RHD-RHCE-RHD gene with a normal RHD exon 10.37 38 It is possible that such a rearranged gene may be present in other individuals expressing the dCe phenotype that are RHD exon 10 positive by PCR assay.
Nucleotide sequence of Rh D transcript isolated from donor 538150 in the codon 41 region. The sequence electropherogram from the Applied Biosystems 373A DNA sequencer is displayed. The single point mutation generating the in-frame stop codon occurs at codon 41. The nucleotide sequence and inferred amino acid sequence is illustrated below the electropherogram.
Nucleotide sequence of Rh D transcript isolated from donor 538150 in the codon 41 region. The sequence electropherogram from the Applied Biosystems 373A DNA sequencer is displayed. The single point mutation generating the in-frame stop codon occurs at codon 41. The nucleotide sequence and inferred amino acid sequence is illustrated below the electropherogram.
The existence of such individuals is problematic when one considers that RHD typing assays for use in prenatal determination were initially based on the tenet that all D− negative chromosomes lack the RHD gene. Our findings and of others17,18,20 indicate that caution should be applied in prenatal determination with the clear understanding that, although rare, false positive results are unavoidable with dCe and dcE haplotypes. Therefore, it is essential that laboratories performing Rh D typing assays use the amplification of at least two different regions of the RHD gene. Intron 4 and exon 10 are sufficiently distant to be good candidate regions. It is also essential that laboratories wishing to perform RHD prenatal diagnosis should consider the ethnic group of the population they will be potentially screening as the genetic basis (or bases) underlying the D− phenotypes in such populations may well be different to that described for the Caucasian dce chromosomes described initially.1 Further investigation of the genetic mechanisms which generate the D− phenotype in individuals of different ethnic groups is necessary.
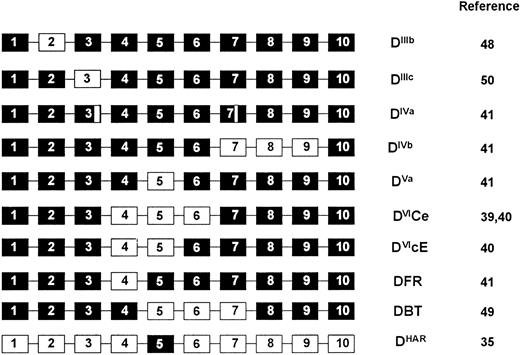
RHD gene organization in D+, partial D, and weak D phenotypes.In individuals expressing normal D+ phenotypes (representing the majority of D+ phenotypes) the multiplex assay is concordant (Table 1). However, in D-variant and weak D phenotypes, the assay highlights many of these individuals would be negative using single Rh D typing assays based on exon 10 or intron 4 of the RHD gene (Tables 1 and 2). In one instance the assay was negative with both intron 4 and exon 10 components in a D-variant (DHar). In the RHaro phenotype, a segmental exchange between the RHCE and RHD genes resulting in essentially a RHCE gene with a RHD exon 5 occurs35 (Fig 7). We also confirm that all RHD genes in individuals expressing the DVI phenotype appear to lack intron 4 of the RHD gene. A hybrid RHD-RHCE-RHD gene, resulting from a probable gene conversion event is known to generate this phenotype39,40 (Fig 7). The multiplex assay could be applied in instances of fetomaternal alloimmunization in mothers expressing the DVI phenotype.40 In two individuals expressing the DFR variant phenotype we show that these appear to occur on two different genetic backgrounds, one individual being RHD intron 4 positive, the other negative (Table 1). As this phenotype appears to arise from a gene conversion where exon 4 of RHD is replaced by RHCE equivalents41 (Fig 7), we assume that intron 4 can be either RHD or RHCE-derived.
Gene backgrounds of partial D phenotypes. D variant phenotypes that are proposed to arise through gene conversion or double cross-over events are shown. The hybrid genes are represented as 10 exons, of which some (black boxes) are derived from the RHD gene, and others (white boxes) from the RHCE gene. The DIVa phenotype involves either point mutation or gene conversion within exons 3 and 7 of the RHD gene, and are thus depicted as small sections of RHCE exons.
Gene backgrounds of partial D phenotypes. D variant phenotypes that are proposed to arise through gene conversion or double cross-over events are shown. The hybrid genes are represented as 10 exons, of which some (black boxes) are derived from the RHD gene, and others (white boxes) from the RHCE gene. The DIVa phenotype involves either point mutation or gene conversion within exons 3 and 7 of the RHD gene, and are thus depicted as small sections of RHCE exons.
Weak D phenotypes are not partial D phenotypes, but represent any D-positive erythrocyte sample with a depression in the apparent numbers of D antigen sites. The molecular basis of the weak D phenotype is poorly defined23,42,43 but has been found to be due to a depression of Rh D mRNA levels in three different individuals.44 Our studies indicate there is genetic heterogeneity in the weak D phenotype individuals examined in this study. It is possible that those weak D phenotype individuals who type as RHD intron 4 negative are uncharacterized partial D phenotypes. However, it is possible to discount the possibility that these are DVI phenotypes, as these erythrocytes typed positive with an anti-D that does not react with these erythrocytes. It is important to identify partial D-phenotype individuals (especially women of child bearing age), as they may be alloimmunized on exposure to normal D+ blood.
The discordance of any PCR-based Rh D typing assay in D+ phenotypes is concerning. However, we show that discordance is confined to weak D and D-variant phenotypes, and there has been only one published example of fetomaternal alloimmunization due to a fetus expressing these phenotypes.47 This is most likely due to the significantly depressed D antigen site numbers on these erythrocytes.45 Alloimmunization has been described in mothers expressing D-variant phenotypes and carrying normal D+ fetuses.46 47 As with D− phenotypes, the use of a multiplex assay minimizes the risk of false negative results that may be obtained when using a single RHD gene assay. It is possible that multiplexing using more than two different regions in the RHD gene may be necessary.
ACKNOWLEDGMENT
We thank Dr G. Galea (Inverness RTC) and Dundee RTC; H. Fogg, Trent RTC, Sheffield; and the serum cells and rare fluids (SCARF ) scheme (distributed by Gamma Biologicals Inc, Houston, TX) for erythrocytes, Susan Cochrane for technical assistance, and to Dr Jeff Jones, Joyce Poole, and Douglas Clark for serological testing. We also thank Dr Marion Scott for critical reading of the manuscript.
Address correspondence to Neil Avent, PhD, International Blood Group Reference Laboratory, Southmead Rd, Southmead, Bristol, BS10 5ND, UK.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal