Abstract
A peculiar feature of rheumatoid arthritis patients is that they carry clonally expanded CD4+ and CD8+ cells in the peripheral blood. While the distortion of the repertoire of CD8+ cells has been ascribed to the increase of CD8+CD57+ large granular lymphocytes, often detected in these patients, the mechanism responsible for the clonal expansion of CD4+ cells remains unexplained. Here, we report that CD4+CD57+ cells, that in healthy individuals represent a small subset of peripheral CD4+ lymphocytes, are significantly expanded in the peripheral blood of a considerable percentage of rheumatoid arthritis patients. Furthermore, the expansion of these lymphocytes appears to correlate with the presence of rheumatoid factor. The molecular analysis of the T-cell receptor variable beta segments expressed by the CD4+CD57+ cells enriched in rheumatoid arthritis patients showed that they use restricted repertoires, that partially overlap with those of their CD4−CD57+ counterpart. The structural feature of the receptor ligand expressed by these cells revealed that their expansion is most likely mediated by strong antigenic pressures. However, since we also found that CD4+CD57+ and CD4−CD57+ cells can share the same clonal specificity, it is likely that their selection is not mediated by conventional major histocompatibility complex restricted mechanisms. Thus, while our data demonstrate that CD4+CD57+ cells play an important role in establishing the imbalance of the CD4+ cell repertoire observed in rheumatoid arthritis patients, they also suggest that these cells have common features with mouse CD4+CD8−NK1.1+/T cells.
THE ETIOLOGY OF rheumatoid arthritis (RA) has been the subject of intense studies but, unfortunately, it remains elusive.1 The presence of CD4+ T cells in the synovial lesions has supported the hypothesis that T-helper cells, possibly responsive to arthritogenic antigens, are important in inducing and sustaining the inflammatory process,2-4 but recent evidence also indicates that the involvement of T cells is a more complex phenomenon than the simple recognition of an arthritogenic antigen in the synovia. In the absence of a candidate-inducing ligand, experimental approaches have been concentrated on defining the molecular diversity of the T-cell populations selectively expanded in the course of the disease. However, these studies yielded conflicting results on whether particular elements of the T-cell receptor (TCR) variable α (TCRAV) or β (TCRBV) chains were enriched at the site of inflammation or on whether there was a consensus pattern in the TCR distribution in the different patients.5-11 Several studies, on the contrary, agree in demonstrating that the TCR repertoires of RA patients are severely skewed with the emergence of dominant clonotypes in the CD8 population.12-14
Since expansions of large granular lymphocytes (LGL), that can be distinguished by the presence of CD57 surface marker,15 are often detected in RA patients,16,17 the distortion of the repertoire in the CD8 population has been ascribed to the presence of these cells in the blood. Little is known, on the contrary, on the frequency, in RA patients, of CD4+CD57+ lymphocytes, a subset of cells that is expanded in some pathologic conditions, such as after renal allograft18 and human immunodeficiency virus (HIV) infection.19 This issue is of particular interest since clonal expansions in RA patients appear to be not selectively restricted to the CD8 compartment, but also to involve CD4 circulating lymphocytes.20 21 These expanded clonotypes that are present, but not preferentially accumulated, in the synovial fluid, persist for several years.
We have recently characterized, at the molecular level, the TCR repertoires of fractionated T-cell populations of patients with concurrent expansions of CD4+CD57+ and CD8+CD57+LGL and we demonstrated that the dominant TCRBV chains expressed by these two populations were strictly oligoclonal and that their expansions were clearly due to antigen selection.22 Considering the different functions and recognition patterns of CD4+ and CD8+ T cells and the clonal expansion of both subsets in RA, we decided to study the frequencies and the molecular properties of CD4−CD57+ and of CD4+CD57+ cells in RA patients. We report that, among the 25 RA patients analyzed, 10 were characterized by the concurrent expansion of CD8+CD57+ and of CD4+CD57+ lymphocytes. The molecular characterization of the TCRBV genes expressed by these cells revealed unexpected features, suggesting that these lymphocytes play an important role in establishing the imbalance of the TCR repertoire observed in RA patients.
PATIENTS AND METHODS
Patients.The 25 RA patients, diagnosed according to the ARA criteria for classification of RA revised in 1987,23 were randomly chosen among individuals observed at Day Hospital of the Clinical Immunology Department of the Spedali Civili di Brescia (Italy). All patients gave their informed consent to the study.
Preparation of lymphocytes subpopulations.Total lymphocytes were isolated from heparinized blood by Ficoll-Hypaque gradient centrifugation. Fresh CD4+CD57+, CD4+CD57−, CD4−CD57+, and CD4−CD57− subpopulations were prepared by using magnetic microspherical beads coated with specific monoclonal antibodies (MoAbs; Dynabeads M-450 CD4 and Dynabeads M-450 rat antimouse IgM; Dynal, Oslo, Norway) and DETACHaBEAD, following the manufacturer's instructions. The morphology of LGL and the purity of cell subsets, analyzed by flow cytometry, were studied after each step of the preparations. Morphologic analysis for evaluation of LGL was performed on May-Grünwald Giemsa–stained cytospin: the LGL were large or medium sized cells, with moderate to abundant pale blue cytoplasm containing azurophilic granules.
Immunofluorescence analysis.A standard cytofluorimetric analysis of peripheral blood lymphocytes was performed using a one-color or two-color whole blood staining method with a panel of purified, phycoerythrin- or fluorescein isothiocyanate-conjugated MoAbs. Anti-CD3, anti-CD4, anti-CD8, anti-CD56, anti-CD57, anti–HLA-DR, anti-TCRAB, anti-CD45RA and anti-CD45RO MoAbs were obtained from Becton Dickinson (Mountain View, CA), while the anti-TCRGD MoAb was from T-cell Sciences (Cambridge, MA). The anti-CD16 and the anti-CD20 MoAbs were purchased from Ortho (Ortho Diagnostic System, Raritan, NJ) and the anti-CD25 MoAb was from Coulter Immunology (Hialeah, FL). Cells were stained according to the manufacturer's instructions. Ten thousand cells were accumulated for histograms using logarithmic amplification of fluorescence intensity on a FACScan flow cytometer (Becton Dickinson, Erembodegem-aalst, Belgium).
Preparation of RNA, cDNA synthesis, and amplification of cDNA by PCR.Total cytoplasmatic RNA and cDNA were prepared from all lymphocyte subpopulations as previously described.22 cDNA was subjected to enzymatic amplification using a TCRBC primer (βAI: 5′ CCC ACT GTG CAC CTC CTT CC 3′ ) and a TCRBV degenerated primer [Vβd: 5′ ACG TGA ATT CT(GT) T(ACT)(CT) TGG TA(CT) (AC)(AG)(AT) CA 3′] that was designed to allow the simultaneous amplification of all the TCRBV segments in a given sample.24 The 40 cycles of PCR were carried out under the following conditions: denaturation at 93°C for 1 minute, annealing at 52°C for 1 minute, and extension at 72°C for 1 minute; the last cycle extension was carried out at 72°C for 7 minutes. The specificity of the total amplified products was analyzed using a colorimetric method and biotinylated TCRBV-specific probes25 and the relative percentage of expression of each of the TCRBV segments analyzed was calculated by normalizing the optical density value (O.D.) of each individual TCRBV segment with respect to the sum of the O.D. values of all the 26 TCRBV chains as follows:
Subsequently, the TCRBV chains of interest were amplified by 35 cycles of PCR22 using TCRBV-specific family primers (TCRBV1: 5′ GCA CAA CAG TTC CCT GAC TTG CAC 3′; TCRBV2: 5′ TCA TCA ACC ATG CAA GCC TGA CCT 3′; TCRBV3: 5′ GTC TCT AGA GAG AAG AAG GAG CGC 3′; TCRBV5S2: 5′ TTC CCT AAC TAT AGC TCT GAG CTG 3′; TCRBV6: 5′ AGG CCT GAG GGA TCC GTC TC 3′; TCRBV13S2 5′ GGT GAG GGT ACA ACT GCC 3′; TCRBV14: 5′ GTC TCT CGA AAA GAG AAG AGG AAT 3′ and TCRBV24: 5′ CCA ATC CAG GAG GCC GAA CAC TTC 3′ ) and the TCRBC oligonucleotide βAI.
Heteroduplex analysis.Twenty microliters of all PCR reactions, performed with TCRBV-specific primers, were heated to 95°C for 5 minutes and cooled to 50°C for 1 hour. Samples, kept on ice until used, were loaded on 12% nondenaturating polyacrylamide gel electrophoresis (PAGE; 29:1 acrylamide/bisacrylamide) performed in TBE buffer (0.089 mol/L Tris-borate and 0.002 mol/L EDTA, pH 8.0). Gels were run for 5 to 6 hours at 200 V, at room temperature. DNA molecular weight V (Boehringer Mannheim GmbH, Mannheim, Germany) was used as size marker. The gels were stained for 30 to 60 minutes, at room temperature, in the dark, in a solution containing 0.75 μg/mL ethidium bromide in 200 mL of TBE. Amplified TCRBV8 products from the T-cell line J77 and from lymphocytes stimulated with an anti-TCRBV8 MoAb, prepared as previously described,26 were used in these experiments, respectively, as monoclonal and polyclonal controls.
Cloning and sequencing of PCR products.The specific PCR products were purified by cutting the band with the expected size from Nu-Sieve GTG low melting agarose 2.5% gel (FMC BioProducts, Rockland, ME) and eluting the melted gel through ion-exchange resin column (Qiagen tip 5; Qiagen Inc, Chatsworth, CA). Purified DNA fragments were ligated to pCR II vector. Plasmids were grown in INVαF′ modified competent Escherichia coli cells in LB agar plates and single plaques were picked up and expanded according to the manufacturer's instructions (TA cloning kit; Invitrogen Corp, San Diego, CA). The presence of the correct insert was verified by PCR with the above described TCRBV-specific family primers and with 2 primers specific for the TCRBC segment (the anti-sense βAI primer and the sense oligonucleotide Cβ1: 5′ GTC GCT GTG TTT GAG CCA TCA GAA 3′ ). Plasmid DNA from TCRBV positive plaques was prepared using Qiawell 8 Plasmid kit (Qiagen) and sequenced with Automated Laser Fluorescent A.L.F. DNA Sequencer (Pharmacia LKB, Uppsala, Sweden) using AutoRead Sequencing Kit (Pharmacia). Sequences were compared with published data relative to TCRBV, TCRBD, TCRBJ, and TCRBC segments.27-33
RESULTS
Identification of CD4+CD57+ lymphocytes in RA patients.In a first series of experiments we investigated the expression of the CD57 marker on CD4+ lymphocytes obtained from 25 randomly chosen RA patients. While CD4+CD57+ cells represent a minor subset of CD4+ T lymphocytes in peripheral blood of normal individuals, we found that they were expressed at high levels in some RA patients (Table 1). On the basis of the presence of circulating CD4+CD57+ cells, the 25 RA patients could be divided into two groups: a first one of 15 patients with a number of CD4+CD57+ cells similar to that of normal individuals (25.3 ± 18.1 v 30.1 ± 23.5) and a second one of 10 patients with an abnormally elevated number of CD4+CD57+ lymphocytes (140.5 ± 10.0 v 30.1 ± 23.5, P < .001). The increased level of CD4+CD57+ cells was not apparently related to changes in the percentage and number of CD4+ cells because they were similar in all groups. In the second group of patients there was, however, a concomitant significant increase in the number of CD3+ lymphocytes bearing the CD57 marker in comparison with the first group and controls (477.8 ± 127.4 v 144.2 ± 86.0, P < .001 and v 158.5 ± 95.1, P < .001). From the analysis of the values of the individual patients belonging to this last group it is also evident that the expansion of CD57+ cells does not necessarily correlate with an increase of CD4+CD57+ or CD8+CD57+ lymphocytes.
Clinical Characteristics and Immunophenotypic Analysis of RA Patients
| Patients . | Group . | Sex . | Age (yrs) . | ARA Class . | RF . | Duration (yrs) . | Therapy . | Lymphocytes (×103) . | CD3+ (%) . | CD4+ (%) . | CD8+ (%) . | CD57+ (%) . | CD3+CD57+ (%) . | CD8+CD57+ (%) . | CD4+CD57+ . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | % . | μL . |
| MAAP | 1 | F | 61 | I | − | 20 | DPA, NSAID, S | 1.4 | 48.5 | 31.2 | 22.7 | 27.4 | 2.9 | 8.7 | 0.3 | 4.2 |
| ROBR | F | 41 | I | + | 12 | CSA, MTX, S | 1.4 | 83.0 | 65.5 | 18.5 | 9.1 | 6.7 | 5.3 | 0.5 | 7.0 | |
| GRMU | F | 31 | II | + | 11 | HCQ, NSAID, S, SSZ | 2.3 | 71.1 | 31.1 | 41.0 | 18.8 | 13.0 | 11.6 | 0.6 | 13.8 | |
| ROGR | F | 48 | II | + | 7 | CSA, MTX, S, NSAID | 2.8 | 74.5 | 53.9 | 22.8 | 11.1 | 5.3 | 6.3 | 0.6 | 16.8 | |
| TEAG | F | 64 | II | − | 14 | HCQ, NSAID, S | 1.6 | 67.5 | 46.8 | 21.2 | 18.8 | 11.9 | 7.7 | 0.7 | 11.2 | |
| OSGA | M | 40 | III | − | 11 | CSA, GST, MTX, NSAID, S | 2.2 | 88.1 | 55.4 | 33.6 | 7.5 | 2.6 | 3.0 | 0.7 | 15.4 | |
| GAAR | F | 63 | III | + | 5 | GST, MTX, NSAID, S | 1.4 | 87.7 | 70.7 | 19.8 | 3.1 | 1.4 | 1.4 | 0.7 | 9.8 | |
| BRCA | M | 39 | III | + | 1 | HCQ, MTX, NSAID, S, SSZ | 1.4 | 72.3 | 45.7 | 36.3 | 8.9 | 7.3 | 7.9 | 1.0 | 14.0 | |
| DAFR | M | 59 | II | − | 13 | GST, NSAID, S | 2.7 | 68.0 | 60.2 | 8.4 | 7.6 | 4.3 | 5.2 | 2.1 | 56.7 | |
| ANAP | M | 48 | II | + | 12 | AF, NSAID | 2.4 | 55.8 | 41.0 | 27.4 | 30.8 | 12.4 | 16.2 | 2.2 | 52.8 | |
| GESA | F | 71 | IV | + | 16 | NSAID, S | 1.4 | 49.7 | 29.8 | 51.3 | 45.1 | 18.2 | 31.1 | 2.2 | 30.8 | |
| CABO | F | 53 | II | − | 21 | NSAID, S | 1.6 | 48.0 | 32.5 | 37.4 | 38.5 | 9.5 | 20.1 | 2.5 | 40.0 | |
| ROPE | F | 56 | III | − | 15 | NSAID, S | 1.1 | 82.5 | 53.1 | 26.8 | 14.0 | 10.2 | 8.6 | 3.0 | 33.0 | |
| PIZI | M | 56 | II | + | 12 | HCQ, MTX, NSAID, S, SSZ | 0.6 | 78.0 | 54.0 | 23.7 | 22.4 | 18.4 | 12.9 | 3.3 | 19.8 | |
| CAZI | F | 52 | III | + | 12 | MTX, NSAID, S, SSZ | 1.4 | 84.1 | 57.2 | 30.8 | 15.8 | 12.2 | 8.6 | 3.9 | 54.6 | |
| MAPI | 2 | M | 64 | III | + | 30 | GST, MTX, NSAID | 2.7 | 85.7 | 63.5 | 26.4 | 16.8 | 14.3 | 13.2 | 4.8 | 129.6 |
| ROPI | F | 54 | III | + | 10 | CTX, GST, MTX, NSAID, S, SSZ | 2.7 | 77.7 | 54.9 | 22.2 | 24.5 | 18.5 | 10.0 | 6.0 | 162.0 | |
| PIPE | F | 69 | III | − | 13 | HCQ, MTX, NSAID, S | 2.5 | 82.1 | 58.7 | 29.1 | 23.3 | 16.7 | 13.4 | 6.1 | 152.5 | |
| GEGU | M | 62 | II | + | 9 | MTX, NSAID, S, SSZ | 1.8 | 74.2 | 46.3 | 32.0 | 38.4 | 26.3 | 22.6 | 7.0 | 126.0 | |
| GIVI | M | 54 | II | + | 2 | HCQ, MTX, NSAID, S, SSZ | 2.3 | 81.3 | 48.7 | 33.0 | 33.8 | 26.7 | 19.3 | 7.1 | 163.3 | |
| GIFE | M | 53 | II | + | 8 | MTX, NSAID, Q, SSZ | 2.0 | 88.0 | 40.4 | 54.9 | 36.1 | 33.8 | 32.6 | 7.2 | 144.0 | |
| INZA | F | 71 | III | + | 18 | DPA, MTX, NSAID, S | 1.5 | 80.4 | 57.6 | 32.9 | 35.0 | 22.4 | 19.0 | 8.3 | 124.5 | |
| LOME | M | 66 | II | + | 5 | NSAID, S, SSZ | 1.2 | 84.8 | 46.7 | 38.9 | 39.7 | 34.2 | 26.8 | 8.4 | 101.0 | |
| VIZA | F | 68 | II | + | 3 | HCQ, MTX, NSAID, S, SSZ | 1.4 | 84.6 | 31.5 | 46.5 | 47.8 | 45.9 | 36.1 | 8.6 | 120.0 | |
| MAGHI | F | 37 | I | + | 7 | HCQ, GST, NSAID | 1.8 | 57.8 | 41.4 | 20.0 | 35.5 | 18.0 | 8.4 | 10.1 | 181.8 | |
| Controls | M/F | 38.8 ± 5.5 | — | — | — | — | 2.0 ± 0.4 | 75.4 ± 8.3 | 50.4 ± 7.3 | 29.3 ± 6.7 | 16.5 ± 7.1 | 8.0 ± 4.6 | 10.4 ± 4.1 | 1.5 ± 1.2 | 30.1 ± 23.5 | |
| Patients . | Group . | Sex . | Age (yrs) . | ARA Class . | RF . | Duration (yrs) . | Therapy . | Lymphocytes (×103) . | CD3+ (%) . | CD4+ (%) . | CD8+ (%) . | CD57+ (%) . | CD3+CD57+ (%) . | CD8+CD57+ (%) . | CD4+CD57+ . | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | % . | μL . |
| MAAP | 1 | F | 61 | I | − | 20 | DPA, NSAID, S | 1.4 | 48.5 | 31.2 | 22.7 | 27.4 | 2.9 | 8.7 | 0.3 | 4.2 |
| ROBR | F | 41 | I | + | 12 | CSA, MTX, S | 1.4 | 83.0 | 65.5 | 18.5 | 9.1 | 6.7 | 5.3 | 0.5 | 7.0 | |
| GRMU | F | 31 | II | + | 11 | HCQ, NSAID, S, SSZ | 2.3 | 71.1 | 31.1 | 41.0 | 18.8 | 13.0 | 11.6 | 0.6 | 13.8 | |
| ROGR | F | 48 | II | + | 7 | CSA, MTX, S, NSAID | 2.8 | 74.5 | 53.9 | 22.8 | 11.1 | 5.3 | 6.3 | 0.6 | 16.8 | |
| TEAG | F | 64 | II | − | 14 | HCQ, NSAID, S | 1.6 | 67.5 | 46.8 | 21.2 | 18.8 | 11.9 | 7.7 | 0.7 | 11.2 | |
| OSGA | M | 40 | III | − | 11 | CSA, GST, MTX, NSAID, S | 2.2 | 88.1 | 55.4 | 33.6 | 7.5 | 2.6 | 3.0 | 0.7 | 15.4 | |
| GAAR | F | 63 | III | + | 5 | GST, MTX, NSAID, S | 1.4 | 87.7 | 70.7 | 19.8 | 3.1 | 1.4 | 1.4 | 0.7 | 9.8 | |
| BRCA | M | 39 | III | + | 1 | HCQ, MTX, NSAID, S, SSZ | 1.4 | 72.3 | 45.7 | 36.3 | 8.9 | 7.3 | 7.9 | 1.0 | 14.0 | |
| DAFR | M | 59 | II | − | 13 | GST, NSAID, S | 2.7 | 68.0 | 60.2 | 8.4 | 7.6 | 4.3 | 5.2 | 2.1 | 56.7 | |
| ANAP | M | 48 | II | + | 12 | AF, NSAID | 2.4 | 55.8 | 41.0 | 27.4 | 30.8 | 12.4 | 16.2 | 2.2 | 52.8 | |
| GESA | F | 71 | IV | + | 16 | NSAID, S | 1.4 | 49.7 | 29.8 | 51.3 | 45.1 | 18.2 | 31.1 | 2.2 | 30.8 | |
| CABO | F | 53 | II | − | 21 | NSAID, S | 1.6 | 48.0 | 32.5 | 37.4 | 38.5 | 9.5 | 20.1 | 2.5 | 40.0 | |
| ROPE | F | 56 | III | − | 15 | NSAID, S | 1.1 | 82.5 | 53.1 | 26.8 | 14.0 | 10.2 | 8.6 | 3.0 | 33.0 | |
| PIZI | M | 56 | II | + | 12 | HCQ, MTX, NSAID, S, SSZ | 0.6 | 78.0 | 54.0 | 23.7 | 22.4 | 18.4 | 12.9 | 3.3 | 19.8 | |
| CAZI | F | 52 | III | + | 12 | MTX, NSAID, S, SSZ | 1.4 | 84.1 | 57.2 | 30.8 | 15.8 | 12.2 | 8.6 | 3.9 | 54.6 | |
| MAPI | 2 | M | 64 | III | + | 30 | GST, MTX, NSAID | 2.7 | 85.7 | 63.5 | 26.4 | 16.8 | 14.3 | 13.2 | 4.8 | 129.6 |
| ROPI | F | 54 | III | + | 10 | CTX, GST, MTX, NSAID, S, SSZ | 2.7 | 77.7 | 54.9 | 22.2 | 24.5 | 18.5 | 10.0 | 6.0 | 162.0 | |
| PIPE | F | 69 | III | − | 13 | HCQ, MTX, NSAID, S | 2.5 | 82.1 | 58.7 | 29.1 | 23.3 | 16.7 | 13.4 | 6.1 | 152.5 | |
| GEGU | M | 62 | II | + | 9 | MTX, NSAID, S, SSZ | 1.8 | 74.2 | 46.3 | 32.0 | 38.4 | 26.3 | 22.6 | 7.0 | 126.0 | |
| GIVI | M | 54 | II | + | 2 | HCQ, MTX, NSAID, S, SSZ | 2.3 | 81.3 | 48.7 | 33.0 | 33.8 | 26.7 | 19.3 | 7.1 | 163.3 | |
| GIFE | M | 53 | II | + | 8 | MTX, NSAID, Q, SSZ | 2.0 | 88.0 | 40.4 | 54.9 | 36.1 | 33.8 | 32.6 | 7.2 | 144.0 | |
| INZA | F | 71 | III | + | 18 | DPA, MTX, NSAID, S | 1.5 | 80.4 | 57.6 | 32.9 | 35.0 | 22.4 | 19.0 | 8.3 | 124.5 | |
| LOME | M | 66 | II | + | 5 | NSAID, S, SSZ | 1.2 | 84.8 | 46.7 | 38.9 | 39.7 | 34.2 | 26.8 | 8.4 | 101.0 | |
| VIZA | F | 68 | II | + | 3 | HCQ, MTX, NSAID, S, SSZ | 1.4 | 84.6 | 31.5 | 46.5 | 47.8 | 45.9 | 36.1 | 8.6 | 120.0 | |
| MAGHI | F | 37 | I | + | 7 | HCQ, GST, NSAID | 1.8 | 57.8 | 41.4 | 20.0 | 35.5 | 18.0 | 8.4 | 10.1 | 181.8 | |
| Controls | M/F | 38.8 ± 5.5 | — | — | — | — | 2.0 ± 0.4 | 75.4 ± 8.3 | 50.4 ± 7.3 | 29.3 ± 6.7 | 16.5 ± 7.1 | 8.0 ± 4.6 | 10.4 ± 4.1 | 1.5 ± 1.2 | 30.1 ± 23.5 | |
Abbreviations: RF, rheumatoid factor; AF, auranofin; CTX, cyclophosphamide; CSA, cyclosporin A; DPA, D-penicillamine; GST, gold sodium thiomalate; HCQ, hydroxychloroquine; NSAID, non-steroid antiinflammatory drugs; MTX, methotrexate; S, steroids; SSZ, sulfasalazine.
The high level of CD4+CD57+ cells was not related to the age or sex of the patients, the ARA classification, the duration of the disease, or the antirheumatic therapy. Interestingly, however, there was a correlation with the presence of rheumatoid factor (RF ) that was present in 9 of the 10 (90%) patients belonging to the second group. On the contrary, only 9 of the 15 (60%) patients belonging to the first group were RF+.
TCRBV repertoires expressed by CD4+CD57+, CD4+CD57−, CD4−CD57+, and CD4−CD57− cell subsets of RA patients.To better understand the significance of the CD4+CD57+ expansions observed in some RA patients, we randomly selected four subjects belonging to the second group for further phenotypic and molecular studies. As shown in Table 2, the vast majority of these patients' T lymphocytes expressed TCRAB molecules and less than 4% of the cells were TCRGD+. The CD2, CD5, and CD20 molecules, as well as the activation markers HLA-DR and CD25, were within the normal range, with the exception of patient MAGHI in which the CD5 marker was lower than in controls. The CD16 and CD56 NK-related antigens were heterogeneously expressed.
Phenotypic Analysis of the Selected Patients
| Patients . | CD2+ . | CD5+ . | CD16+ . | CD20+ . | CD25+ . | CD56+ . | HLA-DR+ . | TCRAB+ . | TCRGD+ . | CD4+CD45RA+ . | CD4+CD45RO+ . | CD8+CD45RA+ . | CD8+CD45RO+ . | CD4+CD25+ . | CD8+CD25+ . | CD4+HLA-DR+ . | CD8+HLA-DR+ . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ROPI | 87.1 | 85.9 | 8.8 | 4.8 | 1.6 | 11.0 | 9.3 | 75.7 | 2.9 | 13.2 | 41.0 | 10.8 | 6.9 | 1.2 | 0.1 | 1.7 | 1.4 |
| GEGU | 82.6 | 93.2 | 11.5 | 3.1 | 3.7 | 15.7 | 7.4 | 71.9 | 0.5 | 24.4 | 29.6 | 21.5 | 7.8 | 1.9 | 0.9 | 1.9 | 3.7 |
| LOME | 89.9 | 79.5 | 3.4 | 3.0 | 1.5 | 0.6 | 9.8 | 79.2 | 3.4 | 16.6 | 26.7 | 26.4 | 11.4 | 0.0 | 0.2 | 3.5 | 4.7 |
| MAGHI | 75.9 | 59.8 | 15.8 | 9.0 | 0.1 | 5.4 | 9.3 | 48.1 | 3.3 | 29.7 | 23.5 | 16.3 | 3.6 | 0.0 | 0.0 | 2.4 | 1.0 |
| Controls | 80 ± 6 | 78 ± 7 | 15 ± 7 | 8 ± 5 | <2 | 14 ± 5 | 12 ± 3 | 74 ± 8 | <2 | 19 ± 8 | 29 ± 10 | 18 ± 9 | 8 ± 3 | <5 | ND | <4 | ND |
| Patients . | CD2+ . | CD5+ . | CD16+ . | CD20+ . | CD25+ . | CD56+ . | HLA-DR+ . | TCRAB+ . | TCRGD+ . | CD4+CD45RA+ . | CD4+CD45RO+ . | CD8+CD45RA+ . | CD8+CD45RO+ . | CD4+CD25+ . | CD8+CD25+ . | CD4+HLA-DR+ . | CD8+HLA-DR+ . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ROPI | 87.1 | 85.9 | 8.8 | 4.8 | 1.6 | 11.0 | 9.3 | 75.7 | 2.9 | 13.2 | 41.0 | 10.8 | 6.9 | 1.2 | 0.1 | 1.7 | 1.4 |
| GEGU | 82.6 | 93.2 | 11.5 | 3.1 | 3.7 | 15.7 | 7.4 | 71.9 | 0.5 | 24.4 | 29.6 | 21.5 | 7.8 | 1.9 | 0.9 | 1.9 | 3.7 |
| LOME | 89.9 | 79.5 | 3.4 | 3.0 | 1.5 | 0.6 | 9.8 | 79.2 | 3.4 | 16.6 | 26.7 | 26.4 | 11.4 | 0.0 | 0.2 | 3.5 | 4.7 |
| MAGHI | 75.9 | 59.8 | 15.8 | 9.0 | 0.1 | 5.4 | 9.3 | 48.1 | 3.3 | 29.7 | 23.5 | 16.3 | 3.6 | 0.0 | 0.0 | 2.4 | 1.0 |
| Controls | 80 ± 6 | 78 ± 7 | 15 ± 7 | 8 ± 5 | <2 | 14 ± 5 | 12 ± 3 | 74 ± 8 | <2 | 19 ± 8 | 29 ± 10 | 18 ± 9 | 8 ± 3 | <5 | ND | <4 | ND |
Values are percentages.
Abbreviation: ND, not done.
To assess whether the expanded CD4+CD57+ T cells shared common features in terms of TCRBV gene usage with other T-cell subsets, we purified by magnetic beads separation the CD4+CD57+, CD4+CD57−, CD4−CD57+, and CD4−CD57− T-cell populations.
Technical impairments, associated with the impossibility of performing a second beads detachment, allowed the assessment of cell purity only in the CD4+CD57− and CD4−CD57− cell subsets. As shown in Table 3, the level of cellular contamination in the CD4+CD57− populations was irrelevant, while we could not avoid the presence of contaminating CD57+ cells in the CD4−CD57− subsets. The cytofluorimetric analysis, however, revealed that a fraction of the contaminating CD57+ lymphocytes appeared to be CD8+CD57+ low positive cells, suggesting that they belong to the pool of lymphocytes that are not positive for the CD3/TCRAB complex.34
Analysis of the T-Cell Subsets' Purity
| Patients . | Cell Populations . | Contaminating Cells (%) . | ||||
|---|---|---|---|---|---|---|
| . | . | CD4+ . | CD8+ . | CD57+ . | CD4+CD57+ . | CD8+CD57+ . |
| ROPI | CD4+CD57− | — | 1.9 | 0.7 | — | 0.2 |
| CD4−CD57− | 6.5 | — | 20.5 | 0.2 | — | |
| GEGU | CD4+CD57− | — | 1.2 | 1.1 | — | 0.2 |
| CD4−CD57− | 5.4 | — | 20.7 | 0.1 | — | |
| LOME | CD4+CD57− | — | 1.9 | 0.5 | — | 0.1 |
| CD4−CD57− | 5.7 | — | 14.7 | 0.1 | — | |
| MAGHI | CD4+CD57− | — | 2.2 | 2.7 | — | 0.3 |
| CD4−CD57− | 1.6 | — | 17.2 | 0.0 | — | |
| Patients . | Cell Populations . | Contaminating Cells (%) . | ||||
|---|---|---|---|---|---|---|
| . | . | CD4+ . | CD8+ . | CD57+ . | CD4+CD57+ . | CD8+CD57+ . |
| ROPI | CD4+CD57− | — | 1.9 | 0.7 | — | 0.2 |
| CD4−CD57− | 6.5 | — | 20.5 | 0.2 | — | |
| GEGU | CD4+CD57− | — | 1.2 | 1.1 | — | 0.2 |
| CD4−CD57− | 5.4 | — | 20.7 | 0.1 | — | |
| LOME | CD4+CD57− | — | 1.9 | 0.5 | — | 0.1 |
| CD4−CD57− | 5.7 | — | 14.7 | 0.1 | — | |
| MAGHI | CD4+CD57− | — | 2.2 | 2.7 | — | 0.3 |
| CD4−CD57− | 1.6 | — | 17.2 | 0.0 | — | |
Since cells bearing the CD57 marker often show LGL morphology,15 we evaluated the presence of LGL at all steps of the samples' preparation. Table 4 shows that the percentage of LGL was elevated in three of the four patients. Globally, the percentage and number of LGL were significantly higher than in normal individuals (24.5% ± 13% v 14.3% ± 2.2%, P = .02 and 510.5 ± 226.5 v 247.6 ± 71.3, P < .02). In the three patients with increased percentage of LGL these cells were preferentially observed in the pool of CD4− lymphocytes. The data relative to the CD4+CD57− and CD4−CD57− preparations allowed us to estimate also the percentage of LGL in the CD4+CD57+ and CD4−CD57+ populations. According with these evaluations, a considerable proportion of CD4+CD57+ cells of patients ROPI and LOME were LGL. This was not, however, the case for patient GEGU, in which LGL represented only 8% of the CD4+CD57+ cells. Thus, CD4+CD57+ cells do not necessarily have the LGL morphology.
Evaluation of the Presence of LGL in the Different T-Cell Subsets
| Patients and Controls . | Lymphocytes (per μL) . | LGL4-150 . | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | PBL . | CD4+ . | CD4− . | CD4+CD57− . | CD4−CD57− . | LGL4-151 . | . | . | . | . | ||||||||||
| . | . | % . | μL . | % . | μL . | % . | μL . | % . | μL . | % . | μL . | CD4+CD57+ . | CD4−CD57+ . | . | . | . | . | . | . | ||
| . | . | . | . | . | . | . | . | . | . | . | . | % . | μL . | % . | μL . | . | . | . | . | . | . |
| ROPI | 2,703 | 24 | 649 | 7 | 104 | 23 | 138 | 0 | 0 | 8 | 26 | 64 | 104 | 41 | 112 | ||||||
| GEGU | 1,861 | 7 | 130 | 2 | 17 | 10 | 60 | 1 | 7 | 4 | 7 | 8 | 10 | 13 | 53 | ||||||
| LOME | 1,251 | 37 | 463 | 11 | 64 | 30 | 146 | 1 | 5 | 7 | 11 | 56 | 59 | 40 | 135 | ||||||
| MAGHI | 1,860 | 30 | 558 | 7 | 54 | 45 | 167 | 0 | 0 | 16 | 35 | 29 | 54 | 85 | 132 | ||||||
| Controls | 2,009 ± 399 | 14.3 ± 2.2 | 247.6 ± 71.3 | 1.2 ± 1.2 | 12 ± 10 | 29 ± 11 | 179 ± 66 | — | — | — | — | — | — | — | — | ||||||
| Patients and Controls . | Lymphocytes (per μL) . | LGL4-150 . | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | . | PBL . | CD4+ . | CD4− . | CD4+CD57− . | CD4−CD57− . | LGL4-151 . | . | . | . | . | ||||||||||
| . | . | % . | μL . | % . | μL . | % . | μL . | % . | μL . | % . | μL . | CD4+CD57+ . | CD4−CD57+ . | . | . | . | . | . | . | ||
| . | . | . | . | . | . | . | . | . | . | . | . | % . | μL . | % . | μL . | . | . | . | . | . | . |
| ROPI | 2,703 | 24 | 649 | 7 | 104 | 23 | 138 | 0 | 0 | 8 | 26 | 64 | 104 | 41 | 112 | ||||||
| GEGU | 1,861 | 7 | 130 | 2 | 17 | 10 | 60 | 1 | 7 | 4 | 7 | 8 | 10 | 13 | 53 | ||||||
| LOME | 1,251 | 37 | 463 | 11 | 64 | 30 | 146 | 1 | 5 | 7 | 11 | 56 | 59 | 40 | 135 | ||||||
| MAGHI | 1,860 | 30 | 558 | 7 | 54 | 45 | 167 | 0 | 0 | 16 | 35 | 29 | 54 | 85 | 132 | ||||||
| Controls | 2,009 ± 399 | 14.3 ± 2.2 | 247.6 ± 71.3 | 1.2 ± 1.2 | 12 ± 10 | 29 ± 11 | 179 ± 66 | — | — | — | — | — | — | — | — | ||||||
The percentage of LGL was evaluated morphologically, while the number of LGL/μL was calculated on the basis of the number of cells with the indicated phenotype.
The number of LGL/μL in the two T-cell subpopulations was deduced by respectively subtracting from the number of LGL found in the CD4+ and CD4− subsets the number of LGL found in the CD4+CD57− and CD4−CD57− cell populations. The percentage of LGL was calculated on the basis of the number of total CD4+CD57+ and CD8+CD57+ circulating lymphocytes.
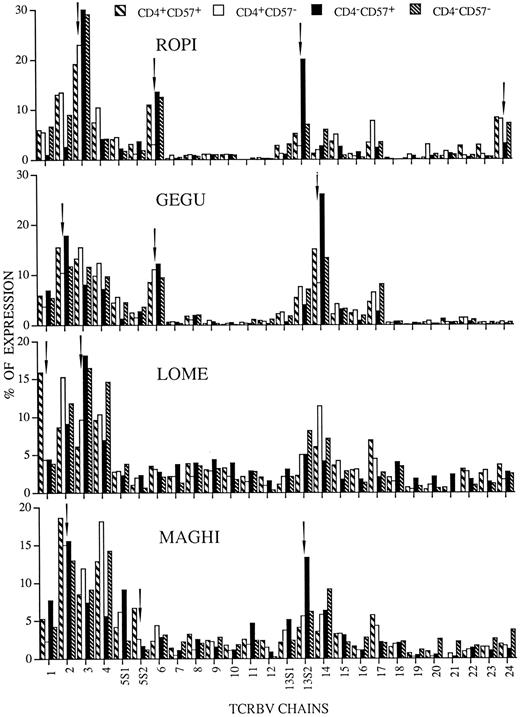
The levels of TCRBV gene usage by the four different T-lymphocyte subsets are shown in Fig 1. The comparative data clearly indicate that a limited number of TCRBV families was overrepresented in some patients, but not in others. Concerning the different T-cell subsets we identified the following patterns of TCRBV expansions: TCRBV segments selectivity enriched in CD4−CD57+ or in CD4+CD57+ cells and TCRBV chains that were overrepresented in both these T-cell subsets. The TCRBV3 segment, for instance, was highly expressed only by CD4−CD57+ lymphocytes of patient LOME, while the TCRBV13S2 chain was dominant only in the CD4−CD57+ subset of patients ROPI and MAGHI. Furthermore, the TCRBV chains overexpressed in both CD4+CD57+ and CD4−CD57+ subpopulations were the following: TCRBV2 segment in patients GEGU and MAGHI, TCRBV3 chain in patient ROPI, TCRBV6 segment in patients ROPI and GEGU, and TCRBV14 chain in patient GEGU. The most unexpected result, however, was the increased expression of TCRBV1 and TCRBV5S2 segments, respectively, observed in CD4+CD57+ lymphocytes of patients LOME and MAGHI. In patient LOME the expansion of TCRBV1 chain was very peculiar, since this gene was only marginally expressed in the other three T-cell subpopulations studied, suggesting the existence of selective pressures acting specifically on CD4+CD57+ cells.
TCRBV expression by CD4+CD57+ (▧), CD4+CD57− (□), CD4−CD57+ (▪), and CD4−CD57− (▧) cell subpopulations prepared from the RA patients ROPI, GEGU, LOME, and MAGHI. The data are expressed as percentage of the colorimetric signal obtained with the individual TCRBV specific probes and each graphic represents the mean of two independent experiments. Arrows indicate the TCRBV families in which the chain is abnormally expressed in at least one of the four T-cell subpopulations.
TCRBV expression by CD4+CD57+ (▧), CD4+CD57− (□), CD4−CD57+ (▪), and CD4−CD57− (▧) cell subpopulations prepared from the RA patients ROPI, GEGU, LOME, and MAGHI. The data are expressed as percentage of the colorimetric signal obtained with the individual TCRBV specific probes and each graphic represents the mean of two independent experiments. Arrows indicate the TCRBV families in which the chain is abnormally expressed in at least one of the four T-cell subpopulations.
Molecular characterization of the dominant TCRBV chains expressed by different T-cell subpopulations.The restriction of the TCRBV repertoires detected in the different T-cell subpopulations of RA patients raised the question of the monoclonal or polyclonal nature of the dominant TCRBV transcripts.
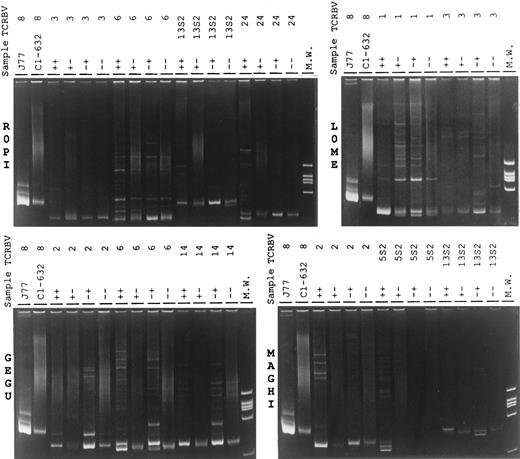
To address this question, we first performed a heteroduplex analysis, that is based on the different ability of amplified TCRBV segments, derived from monoclonal and polyclonal cell preparations, to migrate differently in a polyacrylamide gel. This technique involves the amplification of the target TCRBV chains and the denaturation and renaturation of the amplified product to permit the random association of the distinct DNA strands encoding the different junctional regions. Whereas amplified material from polyclonal lymphoid cells migrates on the polyacrylamide gel as a smear of bands, composed of different sized PCR fragments, the mismatched chains derived from oligoclonal populations migrate as more or less fairly discrete heteroduplex bands of different size that can be separated from the matched homoduplex bands obtained from homogeneous clonal cells. The results, shown in Fig 2, demonstrate that, in all four RA patients, there was a general bias toward mono/oligoclonality of the TCRBV segments derived from CD4+CD57+ and CD4−CD57+ preparations, while the amplified TCRBV segments obtained from CD4+CD57− and CD4−CD57− lymphocytes gave a more polyclonal pattern. The faint homoduplex and heteroduplex bands, often observed with material derived from the CD4−CD57− subpopulations, could be easily ascribed to the documentated contaminations of CD57+ cells. We observed, however, some exceptions to these general pictures. TCRBV3 chain, for instance, gave a polyclonal pattern in all subpopulations of patient ROPI and, at least, in three subsets of patient LOME. TCRBV5S2 segment appeared to be oligoclonal in CD4+CD57+ lymphocytes and undetectable in CD4−CD57+ cells. On the contrary, in patient LOME, the TCRBV1 amplified product derived from the CD4+CD57+ and the CD4−CD57+ subsets migrated, respectively, as a single strong homoduplex band and as a mixture of homoduplexes and heteroduplex bands, superimposed to a polyclonal smear.
Heteroduplex analysis of the indicated TCRBV chains PCR products of different T-lymphocyte subsets prepared from the RA patients ROPI, GEGU, LOME, and MAGHI. PCR products obtained from PCR amplification of the cDNA of the J77 and C1-632 cells27 were used as mono and polyclonal controls. ++, CD4+CD57+ T-cell subset; +− CD4+CD57− T-cell subset; −+, CD4−CD57+ T-cell subset; and − −, CD4−CD57− T-cell subset.
Heteroduplex analysis of the indicated TCRBV chains PCR products of different T-lymphocyte subsets prepared from the RA patients ROPI, GEGU, LOME, and MAGHI. PCR products obtained from PCR amplification of the cDNA of the J77 and C1-632 cells27 were used as mono and polyclonal controls. ++, CD4+CD57+ T-cell subset; +− CD4+CD57− T-cell subset; −+, CD4−CD57+ T-cell subset; and − −, CD4−CD57− T-cell subset.
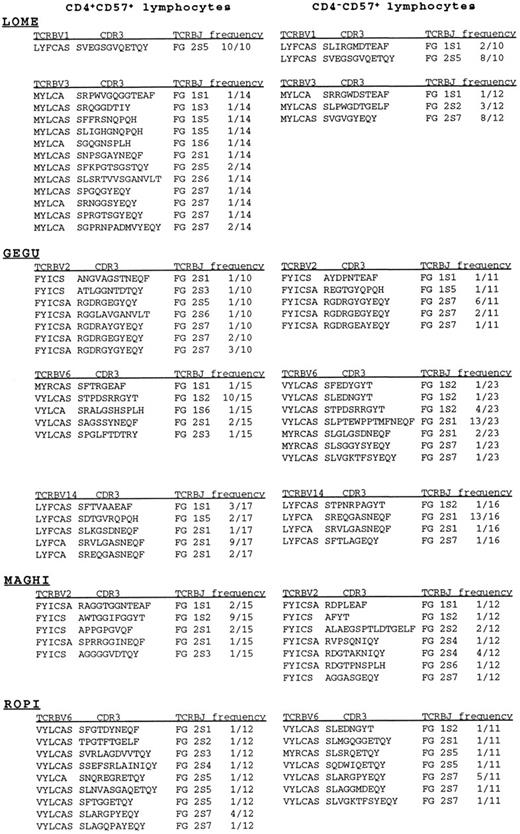
These data were confirmed by the sequencing analysis shown in Fig 3. The repertoire of TCRBV1 chain prepared from CD4+CD57+ lymphocytes of patient LOME was highly restricted and dominated by only one clonal specificity, while the repertoire of the CD4−CD57+ cells was also restricted, but characterized by the presence of at least two different clones. Surprisingly, one of the two dominant transcripts was identical to the dominant one expressed by CD4+CD57+ cells. The possibility that the detection of identical clones in the two T-cell subsets is the result of cell contamination was ruled out by the fact that the dominant TCRBV3 clone, detected in the CD4−CD57+ subset of the same patient, was completely absent in the CD4+CD57+ subpopulation that, on the contrary, was characterized by a polyclonal array of specificities.
Junctional amino acid of TCRBV sequences, obtained from CD4+CD57+ and CD4−CD57+ subpopulations prepared from the RA patients LOME, GEGU, MAGHI, and ROPI were deduced from nucleotide sequences and displayed as standard one-letter code. Only the last 3′ amino acids of the TCRBV segments and the first 5′ amino acids of TCRBJ chains are shown.
Junctional amino acid of TCRBV sequences, obtained from CD4+CD57+ and CD4−CD57+ subpopulations prepared from the RA patients LOME, GEGU, MAGHI, and ROPI were deduced from nucleotide sequences and displayed as standard one-letter code. Only the last 3′ amino acids of the TCRBV segments and the first 5′ amino acids of TCRBJ chains are shown.
Mono/oligoclonal expansions of TCRBV chains were also observed in the two T-cell subsets prepared from patient GEGU. The dominant TCRBV2 transcript was characterized by only one (Y → E and G → A) or two (YG → EA) amino acid substitutions within the CDR3 region. Similarly, the dominant TCRBV14 transcripts found in the CD4+CD57+ and CD4−CD57+ cells differed from each other by only two amino acids (VL → EQ) at the second and third positions of the CDR3 motif. Since, contrary to Ig genes, the TCR variable genes do not undergo the process of somatic mutations, the occurrence of these TCRBV variants is therefore compatible with an ongoing process of antigenic selection, that tends to select those clones with the most closely resembling TCR. The similarity between the dominant clones present in the two cell subsets analyzed is not, however, a general rule since the dominant TCRBV6 chain present in the CD4−CD57+ population of patient GEGU could not be detected in the CD4+CD57+ subset.
Finally, to investigate if recurrent dominant clones are shared by RA patients, we also sequenced the TCRBV2 and TCRBV6 transcripts, obtained, respectively, from patients MAGHI and ROPI, that were found to be mono/oligoclonal at the heteroduplex analysis. TCRBV2 and TCRBV6 cells were found to be more or less restricted in the CD4+CD57+ subset, but we could not find any structural similarity between the dominant clone of these patients and the dominant TCRBV2 and TCRBV6 transcripts found in patient GEGU.
DISCUSSION
It is now well established that RA patients carry clonally expanded CD4+ and CD8+ cells in the peripheral circulation.35 Concerning the mechanism responsible for these clonal expansions, two different models, not mutually exclusive, have been proposed. The first one is based on the hypothesis that clonally expanded T cells may be defective with respect to stimulating- or apoptosis-inducing signals and may therefore escape immunoregulation.36 The second model implies that these T cells may be chronically stimulated and the reason for the clonal TCRBV expansion may be traced to the persistence of an endogenous or exogenous antigen, as it occurs in viral infections such as HIV and Epstein-Barr virus infections.37,38 A few years ago, Legac et al19 demonstrated that the CD4+CD57+ T-cell subset is increased in the peripheral blood of HIV+ patients. Considering that HIV induces a chronic disease, this observation prompted us to investigate whether the CD4+CD57+ cells were also expanded in RA patients. Indeed, in the present work we demonstrate that this particular T-cell population was found to be enriched in a considerable percentage (44%) of the RA patients studied.
The CD57 marker, initially described on large granular CD8+ lymphocytes,39,40 has also been found in a small subset of CD4+ T cells prepared from tonsils of normal individuals.41 The CD57 molecule has also been detected on an important subset of peripheral CD4+ T lymphocytes obtained from renal allograft recipients.18 Interestingly, in these patients, the CD4+CD57+ cells were found to be unresponsive to either lectins or allogeneic cells. More recently, we demonstrated that the dominant TCRBV chains, expressed on CD4+CD57+ and CD4−CD57+ cells from patients with lymphoproliferative disease, were strictly oligoclonal.22 However, the molecular characteristics of the dominant V-D-J rearrangements indicated that these cells had been selected by a strong antigenic pressure.
In our RA patients the increase of CD4+CD57+ lymphocytes did not correlate with the age or sex of the patients, the ARA classification, the duration of the disease, or the antirheumatic therapy. The only significant association with the parameters studied was related to the presence of serum RF. The significance of this correlation is not clear, considering the relatively low number of patients analyzed and the fact that these cell expansions were absent in some patients with RF. It should also be noted that RF specificities in RA patients have been shown to be highly diverse in fine specificity as well as in molecular structure.36 Considering that RF secretion is a T-cell–dependent process, it is possible that CD4+CD57+ cells may provide the stimulus for producing only certain RF type endowed with particular specificities. This possibility and the real significance of the correlation between the expansion of CD4+CD57+ and the presence of RF clearly deserve further studies. It is known, in fact, that RF+ patients have a more severe disease and that extra-articular features of the disease are almost restricted to patients who synthesize autoantibodies with IgG binding reactivity.42
The molecular analysis of the TCRBV segments expressed by the CD4+CD57+ and CD4−CD57+ subsets expanded in RA patients revealed that these T-cell subpopulations use restricted and partially overlapping repertoires. Although it was found that some TCRBV chains were enriched in one subset and not in the other, there were other cases in which the same TCRBV chain was overexpressed in both T-cell subpopulations. Furthermore, our data also establish that the same clonal specificities were present in both CD4+CD57+ and CD4−CD57+ lymphocytes. In the course of the last years at least 370 TCRBV sequences derived from CD4+ and CD4− cells of non-RA individuals. We found that only a very small minority of the sequence derived from CD4+ cells shares the same CDR3 region of the transcript derived from their CD4− counterpart. The finding of identical CDR3 regions between CD4+CD57+ and CD4−CD57+ cells not only confirms and extends our previous results, obtained with two non-RA patients,22 but it also points to the fact that the sharing of the same clonal specificity between CD4 and CD8 cells is not a marginal event but, on the contrary, seems to be a general feature of CD57+ cells. The significance of this phenomenon is even more intriguing considering that the types of rearrangements detected for some TCRBV chains, such as TCRBV14 in patient GEGU, suggest that the selection of these clones results from a strong specific antigenic pressure. The identification of clones using closely resembling, but not identical, TCR structures is, in fact, not compatible with the action of a transforming event leading to a pre-neoplastic condition, that could be the only alternative explanation for the observed clonal expansions.43
The identity, or the similarity, of the receptor ligand expressed by CD4+CD57+ or CD8−CD57+ cells is not compatible with a classical MHC-restricted immune response and therefore must be considered in the light of the nature of the candidate arthritogenic stimuli. It is well known that RA patients have elevated levels of antibodies specific for heat-shock proteins from recombinant bacteria, which are the arthritogenic factors of adjuvant arthritis in rats.44 These evidences have raised new interest on the relation between mycobacteria and rheumatoid arthritis, since animal and bacterial heat-shock proteins have much homology with their human counterpart45 and are believed to have an important role in inflammation. Furthermore, susceptibility to RA is assumed to be carried by the QKRAA motif present in the third hypervariable region of HLA-DRB1*0401.46 Recently, Auger et al demonstrated that this motif specifically binds a 70 kD heat-shock protein.47 In line with these evidences, it has also been reported that in synovial fluid from RA patients there is a large number of CD4−TCRGD+ cells that proliferate in response to mycobacteria antigens.48 Since, in our study, the levels of TCRGD+ cells in the peripheral blood of RA patients were not elevated, it is likely that, if mycobacterial antigens or autologous heat-shock proteins are indeed involved in RA, they must be recognized in the periphery by another subset of specialized cells. We have recently proposed22 that CD4+CD57+LGL+ cells are reminiscent of mouse CD4+CD8−NK1.1+ T cells, a subset of lymphocytes that, contrary to conventional T cells, recognizes proteic, as well as nonproteic antigens, in the contest of CD1 or of other nonclassical MHC class I molecules.49 It has been proposed that the CD1 molecule would preferentially present damaged or inflammatory related molecules such as self heat shock proteins similar to those recognized by some TCRGD+ cells, or a certain class of mycobacterial antigens (eg, mycolic acid) as recently shown in humans.50 Interestingly, all these substances are candidate arthritogenic antigens.44 Thus, NK1.1+ T cells could contribute to nonspecific defense by recognizing damaged cells, or could play a more specific role by recognizing discrete sets of foreign antigens.
Although the existence of the human equivalent of mouse NK1.1+ T cells has not been formally demonstrated, several reports suggest that such cells exist.49 Our findings demonstrating that CD4+CD57+ cells expanded in RA patients possibly in response to arthritogenic stimuli express a restricted TCR repertoire and recognize antigen in a nonclassical MHC-restricted manner, are fully compatible with the hypothesis that these cells represent a specialized subset of T lymphocytes that may be the equivalent of murine CD4+CD8−NK1.1+ T cells. According to this model, the expansion of TCRGD+ cells in the synovia and of CD4+CD57+ in the periphery, possibly mediated by the same ligand, can be most likely explained by the different distribution and modulation of the restricting element (CD1 or nonclassical MHC class I molecule) in the two compartments.
In conclusion, our results have documented that the concurrent expansion of CD8+CD57+ and of CD4+CD57+ lymphocytes with restricted and partially overlapping TCR repertoires is a common event in RA. While our data indicate that these lymphocytes play an important role in establishing the imbalance of the TCR repertoires observed in RA patients, the ultimate proof of the clinical significance of these cells will require an unequivocal assessment of their specificity and function in modulating the disease.
ACKNOWLEDGMENT
We thank the RA patients for participating in the present study and Dr Eugenia Quiros-Roldan for performing initial experiments.
Supported by Sorin Biomedica (Saluggia, Italy) and by Consiglio Nazionale delle Ricerche (Grant No. 96.00710.PF39).
Address reprint requests to Daniele Primi, PhD, Consorzio per le Biotecnologie, Laboratorio di Biotecnologie, P. le Spedali Civili, 1, 25123 Brescia, Italy.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal