Abstract
Previous investigations of exfoliated oropharyngeal cells from individuals suffering from infectious mononucleosis (IM) suggested that the oropharyngeal epithelia are the primary target and also the site of life-long persistence of the Epstein-Barr virus (EBV). This concept was widely accepted. However, the investigation of histological sections with more sensitive EBV detection techniques has drawn this concept into doubt since EBV proved to be constantly absent in normal epithelial cells. To elucidate the discrepancy, throat washings and peripheral mononuclear blood cells from 16 patients suffering from IM were investigated for EBV-DNA and EBV gene products employing highly sensitive in situ hybridization, immunocytochemistry, and polymerase chain reaction. Although all patients exhibited latently infected B lymphocytes in peripheral blood, samples of exfoliated oropharyngeal cells were constantly EBV-negative with the exception of three cases. In these cases, the patients additionally suffered from purulent ulcerating tonsillitis, EBV-infected B cells, but no EBV-infected epithelial cells were detectable. These findings support the view that recirculating lymphocytes of B-cell origin, but not epithelial cells are the initial target of EBV during primary infection and that B cells also represent the site of life-long viral persistence.
THE EPSTEIN-BARR-VIRUS (EBV), an ubiquitous gamma herpes virus, is the etiologic agent of infectious mononucleosis (IM), a self-limiting lymphoproliferative disorder occurring mostly in young adults in association with initial infection.1 IM is characterized by an expansion of infected B lymphocytes, a reactive proliferation of cytotoxic T cells, and oropharyngeal shedding of infectious virions. Following primary infection, EBV establishes a life-long usually asymptomatic carrier-state.2
The role of oropharyngeal epithelium during primary infection is controversial. EBV-specific DNA and RNA have been reported to be present within exfoliated oropharyngeal epithelial cells of patients with acute IM,3,4 which prompted the notion that the epithelium is the primary target of EBV and the cellular site of a primary viral replication with subsequent infection of B lymphocytes. Based on these reports and the constant detection of EBV in epithelial cells of undifferentiated nasopharyngeal carcinoma (UNPC)5 and oral hairy leukoplakia (OHL),6 it has been further concluded that infected epithelial cells are also responsible for the long-term virus persistence.7 This concept has achieved wide acceptance in the last decade. It is, however, challenged by more recent immunohistological and in situ hybridization studies that did not detect EBV in epithelial cells of the oral mucosa in normal and IM tonsils where the presence of EBV proved to be restricted to lymphoid cells.8-11 Furthermore, substantial evidence has been accumulated favoring the view that lymphocytes rather than epithelial cells represent the site of EBV persistence in asymptomatic virus carriers.12-15
As techniques for the demonstration of EBV have greatly improved since the previous studies on EBV in exfoliated oropharyngeal cells were performed, we reinvestigated exfoliated cells obtained from mouth washings and from peripheral blood during acute IM for EBV. For this purpose, we employed a combined approach for the detection of EBV DNA and EBV gene products by the application of polymerase chain reaction (PCR), DNA and RNA in situ hybridization (ISH), and immunocytochemistry (IC). The results obtained show that an EBV infection in exfoliated oropharyngeal cells is only detectable in lymphoid cells of the B-cell lineage, supporting the view that (1) B cells, but not epithelial cells, are the primary target of EBV, and (2) B cells might represent the cellular reservoir responsible for viral persistence.
MATERIALS AND METHODS
Patients and Controls
Throat washings (TW) and heparinized blood samples were obtained from 16 patients with clinical and serological evidence of acute IM between day 3 and 14 after the onset of clinical symptoms. The age range of patients was between 6 and 32 years. Three patients (nos. 6, 9, and 16) also suffered from ulcerating purulent tonsillitis. Six healthy donors served as controls.
Exfoliated cells from the oropharynx were collected by gargling with 15 mL of sterile phosphate-buffered saline (PBS). The collected samples were chilled on ice and processed immediately. Cells were pelleted by low-speed centrifugation and resuspended in PBS, while the supernatant was collected for detection of cell-free viruses. Cytospins were prepared under RNase-free conditions on 3-amino-propyltriethoxysilane (APES)-coated microscope slides, fixed in buffered formalin for 18 hours, dehydrated in graded alcohols, and stored at −80°C until used. Morphological analysis of throat washing cytospin specimens (hematoxylin and eosin staining) showed varying amounts of squamous epithelial cells, granulocytes, monocytes, and lymphocytes between the different cases. Specimens from the majority of donors contained mostly squamous epithelial cells, whereas a predominant leukocyte population was present in the samples of patients with a complicating purulent tonsillitis.
Peripheral blood mononuclear cells (PBMCs) were obtained from blood samples by standard Ficoll gradient centrifugation, washed in PBS, pelleted by low-speed centrifugation, and resuspended in PBS. Cytospins were prepared as described above. DNA was extracted from pelleted and resuspended oropharyngeal cells and PBMCs employing an automated DNA extractor (ABI 341A).
PCR
Two-step nested primer PCR was performed under conditions as described previously16 with modifications of the primers used for reamplification (Table 1). EBV DNA was detected by amplifying part of the BamHI W tandem repeat fragments of EBV. The amplification of part of the β-globin gene served as a control for the extracted DNA amplifiability and was performed in parallel to each EBV amplification. The PCR template consisted of 50 ng DNA each, from both fractions (cells and supernatant of TW), while serial decimal dilutions (100 ng to 10 pg) were used as templates for PBMCs. Amplification of 1 ng of DNA derived from the latently EBV-infected Namalwa cell line (one or two viral copies per cell) served as a positive control. Positive and negative (omission of template DNA) controls were included in every PCR run. Amplification products were examined for bands of appropriate size by ethidium bromide staining and 2% agarose gel electrophoresis. As determined by serial dilutions of genomic DNA from Namalwa cells, the employed PCR assay was capable of detecting a single copy of EBV DNA.
Primers Used for Detection of EBV Genomes in TW and PMBCs Obtained From Patients With IM
| Primer . | Position . | Sequence . |
|---|---|---|
| EBV up | 13980-14000 | 5′-GCAGTAACAGGTAATCTCTGG-3′ |
| EBV low | 14380-14360 | 5′-ACCAGAAATAGCTGCAGGACC-3′ |
| EBV up (R) | 14005-14024 | 5′-GATTTGGACCCGAAATCTGA-3′ |
| EBV low (R) | 14334-14313 | 5′-CCTCCCTAGAACTGACAATTGG-3′ |
| β-globin up | 154-175 | 5′-ATGGTGCACCTGACTCCTGAGG-3′ |
| β-globin low | 508-487 | 5′-GCCATCACTAAAGGCACCGAGC-3′ |
| β-globin up (R) | 212-231 | 5′-ACGTGGATGAAGTTGGTGGT-3′ |
| β-globin low (R) | 462-443 | 5′-TTAGGGTTGCCCATAACAGC-3′ |
| Primer . | Position . | Sequence . |
|---|---|---|
| EBV up | 13980-14000 | 5′-GCAGTAACAGGTAATCTCTGG-3′ |
| EBV low | 14380-14360 | 5′-ACCAGAAATAGCTGCAGGACC-3′ |
| EBV up (R) | 14005-14024 | 5′-GATTTGGACCCGAAATCTGA-3′ |
| EBV low (R) | 14334-14313 | 5′-CCTCCCTAGAACTGACAATTGG-3′ |
| β-globin up | 154-175 | 5′-ATGGTGCACCTGACTCCTGAGG-3′ |
| β-globin low | 508-487 | 5′-GCCATCACTAAAGGCACCGAGC-3′ |
| β-globin up (R) | 212-231 | 5′-ACGTGGATGAAGTTGGTGGT-3′ |
| β-globin low (R) | 462-443 | 5′-TTAGGGTTGCCCATAACAGC-3′ |
Abbreviation: R, reamplification primers.
IC
Immunocytochemistry was performed using a standard alkaline phosphatase antialkaline phosphatase (APAAP) staining procedure and hematoxylin counterstain (Table 2). For double labeling studies, the protocol was modified as described17 to preserve cellular RNA. Productively infected B95-8 cells, IM tonsils, and an oral hairy leukoplakia (OHL) biopsy served as positive controls for IC with the anti-ZEBRA (BZ.1) antibody.
Antibodies Used for Immunochemistry and Lineage Allocation of EBV-Infected Cells in TW and PMBCs Obtained From Patients With IM
| Antibody Designation . | Cluster of Differentiation . | Specificity . | Source . |
|---|---|---|---|
| βF1* | T-cell receptor β chain | T-Cell Sciences (Cambridge, MA) | |
| CD3* (polyclonal) | CD3 | Pan T cell | DAKO (Glostrup, Denmark) |
| CD8* | CD8 | T-cell subset | DAKO |
| L26* | CD20 | Pan B cell | DAKO |
| Ki-B3 | CD45RA | B-cell subset, including certain plasmablasts and proplasma cells | Lab. of Dr. Parwaresch (Kiel, Germany) |
| BZ.1† | EBV-encoded lytic phase BamHI Z (BZLF1) trans-activator protein (ZEBRA) | DAKO |
| Antibody Designation . | Cluster of Differentiation . | Specificity . | Source . |
|---|---|---|---|
| βF1* | T-cell receptor β chain | T-Cell Sciences (Cambridge, MA) | |
| CD3* (polyclonal) | CD3 | Pan T cell | DAKO (Glostrup, Denmark) |
| CD8* | CD8 | T-cell subset | DAKO |
| L26* | CD20 | Pan B cell | DAKO |
| Ki-B3 | CD45RA | B-cell subset, including certain plasmablasts and proplasma cells | Lab. of Dr. Parwaresch (Kiel, Germany) |
| BZ.1† | EBV-encoded lytic phase BamHI Z (BZLF1) trans-activator protein (ZEBRA) | DAKO |
Used only for labeling of PBMCs.
Used only for labeling of TW.
RNA In Situ Hybridization (EBER-ISH)
To identify latently EBV-infected cells in TW and PBMCs, RNA-ISH was performed with radiolabeled RNA probes specific for the small EBV-encoded nuclear transcripts (EBER1 and 2) as described previously.18 EBER-ISH was combined with IC to identify phenotypes of EBER positive cells.
DNA In Situ Hybridization (DNA-ISH)
A highly sensitive, nonisotopic EBV DNA-ISH was applied for detection of EBV genomes in cells from TW to identify productively infected cells lacking EBER expression. Probe DNA for ISH derived from cosmid clones containing the entire EBV genome of the nondefective M-ABA strain stems from nasopharyngeal carcinoma cells.19 Probes were prepared by nick translation with DIG-11–dUTP (Boehringer, Mannheim, Germany) according to the manufacturer's protocol and examined with standard agarose gel electrophoresis and dot blots. Prehybridization was performed as in the protocol used for RNA-ISH. The hybridization mixture contained 50% deionized formamide, 10% dextran sulphate, 2× SSC, 200 μg sonicated salmon testes DNA/mL, and 2 μg labeled probe DNA/mL. A total of 25 μL of hybridization cocktail was applied to each slide, sealed by cover slips, and transferred to a heating block at 90°C for 7 minutes. Hybridization was performed at 37°C overnight in a moist chamber. Cover slips were removed in 2× SSC, washed twice in 20% formamide/0.1× SSC at 42°C for 30 minutes each, and in 2× SSC (42°C and room temperature, respectively) for 15 minutes each. Immunocytological detection of bound probes was performed with the streptavidin-biotin–complex conjugated with alkaline phosphatase (DAKO, Glostrup, Denmark) according to the manufacturer's protocol.
Sensitivity and specificity of this assay was determined by hybridization of several EBV-positive and negative cell lines and paraffin embedded specimens. No signal was obtained with the EBV-negative lymphoid cell line L428 and EBER-negative tonsils. In contrast, hybridization of the productively infected lymphoblastoid cell line B95-8 produced signals ranging from a few spots located in the nuclei to strong labeling of nuclei and cytoplasm. Two EBV copies per cell in latently infected Namalwa cells and approximately 60 EBV copies in Raji cells, respectively, could also be detected, as well as infected lymphocytes in paraffin-embedded specimens from tonsils of patients with acute IM. Very strong signals were detected within the virus producing epithelium of an OHL biopsy.
RESULTS
Technical Approach
Different methods were applied for the analysis of EBV DNA and EBV gene products in PBMCs and TW from IM patients and healthy donors. EBV PCR was employed to demonstrate the presence of viral DNA at maximum sensitivity. For morphological identification of EBV-infected cells, DNA and RNA (EBER) in situ hybridization as well as immunocytochemistry for the detection of BamHI Z-fragment–encoded Epstein-Barr replication activator (ZEBRA) was applied. To clarify the cellular origin of the EBV-infected cells, the in situ hybridization assays were combined with immunocytochemistry (double labeling).
PCR
PCR analysis of TW showed EBV DNA in the supernatant (TW/s), as well in the pelleted oropharyngeal cells (TW/c) in 14 of 15 patients (Table 3). In 11 cases, PCR products could be detected only after reamplification, indicating a low copy number of EBV genomes. In contrast, investigation of samples from patients no. 6, 9, and 16 whose disease was complicated by an ulcerating, purulent tonsillitis showed a much higher amount of EBV in the exfoliated oropharyngeal cells, exhibited by a specific PCR product that was detectable after only one round of amplification.
Results of PCR for Detection of EBV in TW and PMBCs of IM Patients and Controls
| Patient No. . | Age . | Day No. . | PCR TW/s . | PCR TW/c . | PCR PBMC . |
|---|---|---|---|---|---|
| 1 | 17 | 10 | + | + | + |
| 2 | 19 | 5 | + | + | + |
| 3 | 25 | 14 | + | + | + |
| 4 | 7 | 10 | + | + | + |
| 5 | 32 | 11 | + | + | + |
| 6 | 19 | 10 | + | + | + |
| 7 | 13 | 6 | ND | ND | + |
| 8 | 19 | 7 | + | + | + |
| 9 | 18 | 6 | + | + | + |
| 10 | 12 | 4 | − | − | + |
| 11 | 6 | 7 | + | + | + |
| 12 | 26 | 6 | + | + | + |
| 13 | 18 | 10 | + | + | + |
| 14 | 6 | 3 | + | + | + |
| 15 | 15 | 9 | + | + | + |
| 16 | 14 | 5 | + | + | + |
| A | 25 | 0 | − | +3-150 | − |
| B | 37 | 0 | − | − | − |
| C | 40 | 0 | − | − | − |
| D | 19 | 0 | − | − | − |
| E | 28 | 0 | − | − | − |
| F | 25 | 0 | − | − | +3-150 |
| Patient No. . | Age . | Day No. . | PCR TW/s . | PCR TW/c . | PCR PBMC . |
|---|---|---|---|---|---|
| 1 | 17 | 10 | + | + | + |
| 2 | 19 | 5 | + | + | + |
| 3 | 25 | 14 | + | + | + |
| 4 | 7 | 10 | + | + | + |
| 5 | 32 | 11 | + | + | + |
| 6 | 19 | 10 | + | + | + |
| 7 | 13 | 6 | ND | ND | + |
| 8 | 19 | 7 | + | + | + |
| 9 | 18 | 6 | + | + | + |
| 10 | 12 | 4 | − | − | + |
| 11 | 6 | 7 | + | + | + |
| 12 | 26 | 6 | + | + | + |
| 13 | 18 | 10 | + | + | + |
| 14 | 6 | 3 | + | + | + |
| 15 | 15 | 9 | + | + | + |
| 16 | 14 | 5 | + | + | + |
| A | 25 | 0 | − | +3-150 | − |
| B | 37 | 0 | − | − | − |
| C | 40 | 0 | − | − | − |
| D | 19 | 0 | − | − | − |
| E | 28 | 0 | − | − | − |
| F | 25 | 0 | − | − | +3-150 |
Abbreviations: Day no., days since onset of clinical symptoms; A-F, controls; TW/s, supernatant of TW; TW/c, pelleted cells of TW; PBMCs, peripheral blood mononuclear cells; ND, not done.
Detectable only with 100 ng genomic DNA template equivalent to approximately 14,000 cells.
In all IM cases, EBV DNA was detectable by PCR in 1 ng or less of genomic DNA samples extracted from PBMCs. In the control group consisting of six healthy individuals, EBV-specific signals could only be detected when 100 ng genomic DNA was used as a template in the pelleted oropharyngeal cells of one donor and in the PBMCs of another (Table 3).
IC and ISH
PBMCs.Abundant EBER signals could be detected in many PBMCs of all IM patients investigated. As shown by double labeling experiments, most of these cells expressed the CD45RA (Ki-B3 clone)20 antigen, whereas only a minority was CD20 positive (Fig 1A and B). None of the EBER positive PBMCs displayed a T-cell phenotype indicated by the constant absence of CD3, CD8, and the T-cell receptor β chain (Table 4).
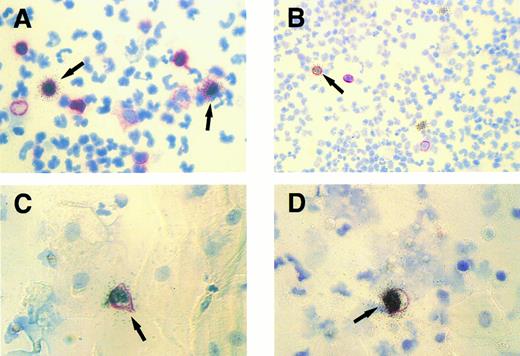
Cytospin specimens of PBMCs and exfoliated oropharyngeal cells from a human affected by IM. Isotopic EBER ISH (silver grains) was combined with immunocytochemistry (red membrane staining). (A) Two EBER- positive peripheral blood lymphocytes (arrows) expressing the CD45RA antigen detected by MoAb Ki-B3. Original magnification × 630. (B) A single EBER-positive peripheral B lymphocyte (arrow) expressing the CD20 antigen (MoAb L26) and two EBER-positive PBMCs lacking CD20 expression. Original magnification × 250. (C and D) Simultaneous labeling for EBER and CD45RA shows single EBV-infected lymphocytes (arrows) among multiple unlabeled epithelial cells characterized by their broad cytoplasm. Original magnification × 630.
Cytospin specimens of PBMCs and exfoliated oropharyngeal cells from a human affected by IM. Isotopic EBER ISH (silver grains) was combined with immunocytochemistry (red membrane staining). (A) Two EBER- positive peripheral blood lymphocytes (arrows) expressing the CD45RA antigen detected by MoAb Ki-B3. Original magnification × 630. (B) A single EBER-positive peripheral B lymphocyte (arrow) expressing the CD20 antigen (MoAb L26) and two EBER-positive PBMCs lacking CD20 expression. Original magnification × 250. (C and D) Simultaneous labeling for EBER and CD45RA shows single EBV-infected lymphocytes (arrows) among multiple unlabeled epithelial cells characterized by their broad cytoplasm. Original magnification × 630.
Immunophenotype of the EBV-Infected Cells Present in TW and PB of IM Patients
| Patient . | PBMC EBER . | PBMC EBER + β-F1 . | PBMC EBER + CD3 . | PBMC EBER + CD8 . | PBMC EBER + CD20 . | PBMC EBER + CD45 RA . | TW/c EBER . | TW/c EBER + CD45 RA . | TW/c ZEBRA . | TW/c DNA-ISH . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | − | ND | ND | + | + | − | − | − | − |
| 2 | + | − | ND | ND | + | + | − | − | − | − |
| 6 | + | − | − | − | + | + | + | + | − | − |
| 7 | + | − | − | − | + | + | ND | ND | ND | ND |
| 8 | + | − | − | − | + | + | − | − | − | − |
| 9 | + | − | − | − | + | + | + | + | − | − |
| 10 | + | − | − | − | + | + | − | − | − | − |
| 11 | + | − | − | − | + | + | − | − | − | − |
| 12 | + | − | − | − | + | + | − | − | − | − |
| 13 | + | − | − | − | + | + | − | − | − | − |
| 14 | ND | ND | ND | ND | ND | ND | − | − | ND | − |
| 15 | + | − | − | − | + | + | − | − | − | − |
| 16 | + | − | − | − | + | + | + | + | − | − |
| Patient . | PBMC EBER . | PBMC EBER + β-F1 . | PBMC EBER + CD3 . | PBMC EBER + CD8 . | PBMC EBER + CD20 . | PBMC EBER + CD45 RA . | TW/c EBER . | TW/c EBER + CD45 RA . | TW/c ZEBRA . | TW/c DNA-ISH . |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | + | − | ND | ND | + | + | − | − | − | − |
| 2 | + | − | ND | ND | + | + | − | − | − | − |
| 6 | + | − | − | − | + | + | + | + | − | − |
| 7 | + | − | − | − | + | + | ND | ND | ND | ND |
| 8 | + | − | − | − | + | + | − | − | − | − |
| 9 | + | − | − | − | + | + | + | + | − | − |
| 10 | + | − | − | − | + | + | − | − | − | − |
| 11 | + | − | − | − | + | + | − | − | − | − |
| 12 | + | − | − | − | + | + | − | − | − | − |
| 13 | + | − | − | − | + | + | − | − | − | − |
| 14 | ND | ND | ND | ND | ND | ND | − | − | ND | − |
| 15 | + | − | − | − | + | + | − | − | − | − |
| 16 | + | − | − | − | + | + | + | + | − | − |
Abbreviation: ND, not done.
TW.Only in specimens of IM patients with complicating purulent tonsillitis (three of 15 cases analyzed), EBER-ISH showed single latently infected cells in the cytospins of TW (Table 4). No EBER-positive cells were detectable in the remaining cases, as well as in healthy donors. All EBER-positive cells corresponded morphologically to lymphocytes and could be colabeled with the B-cell characteristic monoclonal antibody (MoAb) Ki-B3 (CD45RA)20 (Fig 1C and D). In all IM cases and controls analyzed, no infected cells could be detected using the antibody BZ.1 directed against the BamHI Z (BZLF1) replication activator ZEBRA.21 To ensure that productively infected lymphocytes and/or epithelial cells did not escape detection, DNA-ISH was performed with a probe comprising the whole EBV genome. The labeling results obtained with this probe provided identical information as the immunocytochemistry for ZEBRA.
DISCUSSION
IM represents a disease model for studying the target cells of EBV, as the clinical symptoms and serological findings allow rapid identification of individuals undergoing primary infection. For this purpose, several investigators in the 1970s and 1980s studied peripheral blood and exfoliated oropharyngeal cells from TW of IM patients for the presence of EBV. Investigations of the atypical lymphocytosis in peripheral blood of IM patients disclosed that circulating B lymphocytes are infected by the virus resulting in a reactive proliferation of immunologically activated T cells. In TW however, EBV was demonstrated in abundant epithelial, but not in lymphoid cells. Primarily based on these observations, a widely accepted concept has been established proposing that oropharyngeal epithelial cells represent the primary target for EBV infection. According to this concept, EBV should persist and replicate in epithelial cells leading to a secondary infection of adjacent B lymphocytes.3,7 This concept has been challenged by more recent investigations on tissue sections of various oropharyngeal tissue samples from IM patients and healthy individuals because an EBV presence within epithelial cells was not demonstrable.8-10 Because studies of paraffin embedded material might be less sensitive or representative than the analysis of exfoliated oropharyngeal cells, a reevaluation of this discrepancy appeared to be imperative. A reanalysis of exfoliated oropharyngeal cells also appeared to be indicated because the methods now available for the detection of an EBV infection are more sensitive and more reliable than those applied in the past. In this study, we investigated exfoliated oropharyngeal cells and PBMCs of 16 individuals suffering from acute IM, using a combination of highly sensitive PCR, in situ hybridization and double labeling techniques not employed in earlier studies on this topic.
Analysis of peripheral blood of all individuals with clinical and serological evidence of acute IM by PCR verified the expected presence of EBV DNA, confirming the diagnosis of IM. EBV-specific amplificates were also detectable in the peripheral blood from one of the healthy individuals of the control group. The latter finding reflects the situation observed in peripheral blood of asymptomatic EBV carriers, where approximately one EBV-harboring B cell occurs among 106 virus-free lymphocytes leading to an occasional EBV-specific signal in PCR analysis of these samples. The PCR data fully corresponded to the EBER-ISH findings, which showed several EBV infected cells with the morphology of lymphocytes in all samples from IM patients, but not in any of the control group. The combination of EBER-ISH with immunocytochemistry confirmed that the EBER positive lymphocytes belonged to the B-cell lineage, as they were found to express CD45RA (detected by MoAb Ki-B3) and CD20 (in few cells), whereas all T-cell–specific markers (CD3, CD8, and β chain of T-cell receptor) were absent. Decoration of B cells with Ki-B3 combined with the partial absence of CD20 expression is indicative of their plasmacellular differentiation. This finding of plasmacellular differentiated EBER-positive lymphocytes corresponds with previous reports showing that during acute IM up to 80% of EBV nuclear antigen-positive cells in peripheral blood may express cytoplasmic immunoglobulins.22
TW from IM patients were primarily subjected to PCR for EBV detection. EBV-specific amplificates were found in cell-free supernatants, as well as in the exfoliated cell fractions in 14 of 16 IM patients. In samples of three cases, specific amplificates were readily detectable after the first round of amplification, indicating the presence of abundant EBV genomes. Correlation with the clinical findings disclosed that these particular individuals displayed extensive purulent ulcerations of the palatine tonsils. Interestingly, ISH for EBER detection showed positive cells with the morphology of lymphocytes only in these three cases. To identify their lineage allocation, combined IC and EBER-ISH was performed showing that all EBER-positive lymphocytes were also decorated by the CD45RA antibody Ki-B3. In this respect, these lymphocyte cells appear to be identical with those observed in tissue sections of tonsils from IM patients that also showed morphological characteristics of plasmacellular differentiation.10 The detection of EBV-infected B lymphocytes showing plasmacellular differentiation could be an indication that EBV has an affinity for lymphocytes in that differentiation stage. Alternatively, plasmacellular differentiation may be secondary to EBV infection. The latter hypothesis is supported by the finding that EBV infection can induce in vitro polyclonal immunoglobulin synthesis in B cells.23 24
The unexpected detection of the exclusive presence of EBER transcripts in exfoliated lymphocytes and not in epithelial cells raised the question as to whether EBER-ISH was capable of detecting all types of EBV infection in the samples investigated. It has been shown that productively infected cells can escape detection when investigated with EBER-specific probes alone since expression of EBER may be totally downregulated during the phase of replicative infection.25 26 To exclude this possibility, cytospins from all TW (IM and healthy individuals) were immunocytochemically stained for ZEBRA, an EBV encoded replication transactivator. No positive cells could be identified. In addition, DNA-ISH was performed with probes encompassing the whole EBV genome on all samples from IM-suffering and control individuals. Although this probe allows identification of productively infected cells with a high sensitivity (less than 60 virions per cell) regardless of EBV gene expression (eg, EBER or protein expression), no virus producing cells could be detected.
Of course, viral shedding during IM, which could be demonstrated in 14 of our 16 IM cases by PCR detection of EBV-specific DNA in the cell-free fraction of the TW, requires a productive EBV infection. Our inability to visualize the lytically infected cells within exfoliated oropharyngeal cells is, however, not in conflict with our previous observations in tissue sections of tonsils from IM-suffering individuals,10 considering that intraepithelial, productively infected lymphocytes proved to be very rare in comparison to latently infected cells in these specimens. Most of them are encapsulated within the layers of the epithelium, making their release into the oral cavity a rare event. The release of infected B cells appears to be significantly promoted by ulceration. Therefore, it is conceivable that virus producing cells were not detectable in cytospins of TW in the present study, as the total number of EBV-infected cells was too low to include the productively infected B cells that seldom occur.
Our results are in sharp contrast with reports claiming that latent or lytic EBV infection is abundantly present in exfoliated oropharyngeal epithelia of IM patients. The concept that EBV infects primarily epithelial cells of the oral mucosa also does not fit to the observation that epitheliotropism of EBV has been reproducibly shown to be restricted to a few conditions (for review see Niedobitek and Young2 ): (1) latent infection is constantly found only in certain epithelial neoplasms, mainly undifferentiated nasopharyngeal carcinoma (UNPC) where EBV infection is clonal and restricted to the malignant cells27-29 and (2) viral replication without a detectable latent phase occurs only in oral hairy leukoplakia (OHL), a benign lesion of the tongue associated with acquired immunodeficiency syndrome (AIDS).25,26 Several studies by others and our group, however, have confirmed a constant absence of EBV in normal epithelial cells of immunocompetent individuals.8-11,13 That oropharyngeal epithelia are unlikely to represent the common viral reservoir is further underscored by the finding that EBV can be eradicated by bone marrow transplantation with EBV-negative grafts.14 Taken together, the data presented strongly suggest that recirculating B lymphocytes are the real site where EBV persists for a lifetime. This evidence is further supported by the recent findings that (1) in seropositive individuals, EBV is only detectable in circulating CD19 positive B cells,12 and (2) that in virus carriers, latently and productively infected intraepithelial lymphocytes are demonstrable within the nasopharyngeal mucosa.13
Our findings supply a missing link in the chain of evidence arguing against the currently accepted model of EBV infection in which epithelial cells are claimed to be the primary target and the site for long-term persistence of EBV. Although it cannot be excluded that epithelial cells are transitionally infected before the EBV infection becomes clinically manifested, the data provided by our and other investigations strongly favor the scenario that EBV infects B cells directly and not secondarily via replicatively infected epithelial cells and that EBV persists in recirculating B cells that escape the immune surveillance by downregulating the expression of target molecules.
Esterase ectoenzyme on the cell surface of a peripheral blood monocyte from a patient with mycosis fungoides. (Original magnification × 14,500.) (Courtesy of Ann M. Dvorak, MD, Department of Pathology, Beth Israel Hospital, Harvard Medical School, 330 Brookline Ave, Boston, MA 02215.)
Esterase ectoenzyme on the cell surface of a peripheral blood monocyte from a patient with mycosis fungoides. (Original magnification × 14,500.) (Courtesy of Ann M. Dvorak, MD, Department of Pathology, Beth Israel Hospital, Harvard Medical School, 330 Brookline Ave, Boston, MA 02215.)
ACKNOWLEDGMENT
We are grateful to Conny Kreschel and H.-H. Müller for their excellent technical assistance and to Dr H.J. Delecluse for help with the preparation of DNA probes. Dr G.W. Bornkamm, Munich, Germany, kindly provided the M-ABA cosmid clones. We are further indebted to the following physicians and their colleagues for valuable support: Dr R. Baumgarten, Prenzlauer Berg Hospital, Berlin, Germany; Professor Dr H.-R. Nitze, Neukölln Hospital, Berlin, Germany; and Dr S. Völger, St. Joseph Hospital, Berlin, Germany. We would also like to thank Janet Yates for help with the preparation of the manuscript.
This report is part of the doctoral thesis of M.A.K.
Address reprint requests to M. Hummel, PhD, Institute of Pathology, Klinikum Benjamin Franklin, Free University Berlin, Hindenburgdamm 30, 12200 Berlin, Germany.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal