To the Editor:
Among the questions to be answered regarding umbilical cord blood (CB) cell use for bone marrow reconstitution, one concerns the level of contamination with maternal lymphocytes that could be responsible for severe graft-versus-host disease (GVHD) in immunosuppressed recipients.1 2
In the present study, we report data concerning the contamination with maternal cells in 50 consecutive CB units; to this purpose, we used polymerase chain reaction (PCR) amplification of minisatellite sequences followed by chemiluminescent detection.
CB was collected after delivery of term newborns as previously described.3 Genomic DNA was extracted with the salting out method4 from 500 μL of CB and mother's EDTA whole blood. Samples were handled in the preamplification area in a laminar vertical air flow hood using dedicated positive displacement pipettes.
Apolipoprotein B gene (ApoB)5 and D1S806 minisatellite analysis was performed.
Firstly, we performed a PCR assay of all samples using primers that amplify the VNTR 3′ of the ApoB gene. The VNTR was considered to be not informative when the mother was homozygous or had the same alleles as the child. When ApoB was not informative, we amplified the D1S80 polymorphic locus.
Each PCR reaction was performed in 50 μL final volume containing 100 ng of genomic DNA, 50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 9.0, 1.5 mmol/L MgCl2 , 0.1% Triton X-100, 0.15 μmol/L (for ApoB) or 0.4 μmol/L (for D1S80) of each primer, 36 μmol/L of dCTP, 100 μmol/L of each dNTP (of a mix containing dATP, dGTP, and dTTP), 60 μmol/L of biotin-labeled dCTP, and 3 U of Taq DNA polymerase. PCR conditions and primer sequences were those described elsewhere for ApoB5 and for D1S80.6
Seven microliters of PCR product was loaded on a 3% agarose gel and electrophoresed for 16 hours with 1× TBE at 7 V/cm at 4°C.
To assess the sensitivity of our test and to determine the magnitude of the contamination with maternal cells in CB samples, standards obtained mixing 50 ng/μL of DNA from two individuals with alleles of different sizes for each VNTR polymorphism (ApoB, D1S80) at the dilutions 1:500, 1:1,000, and 1:2,500 were amplified in parallel with the other samples and loaded on the gel. After electrophoresis, DNA was transferred from the gel into a Tropilon plus positively charged nylon membrane (Tropix, Bedford, MA) by Southern blotting.
DNA fragments generated in the PCR reaction were labeled through the incorporation of biotinylated dCTP. The detection of PCR products was obtained through two steps: (1) the conjugation of streptavidin-alkaline phosphatase conjugate (Avidix-AP; Tropix) to the biotin-labeled PCR products; and (2) the addition of CSPDR substrate (Tropix), which decomposed upon enzymatic dephosphorylation. The light emission, from the enzyme activated CSPDR substrate resulted in a DNA band pattern that was imaged on x-ray film (Kodak X-Omat AR; Eastman Kodak Co, Rochester, NY).
For the detection of PCR products, the CSPDR protocol Southern-Light Nucleic Acid Detection Systems (Tropix) was followed.7 8
Exposure to x-ray film ranged from 15 minutes to 2 hours, depending on the visualization of the contaminating band in the standard diluted 1:2,500. Every test was performed twice to confirm the results.
For the 50 cases examined, ApoB was informative for 21 samples, whereas for the remaining samples D1S80 was useful.
Noninherited maternal allele was detected in 5 of the 21 CB samples examined with ApoB; with D1S80, 5 additional samples were found contaminated of the 29 informative.
The level of contamination, defined comparing the intensity of the contaminating band with that of the standards, ranged from 1:100 to 1:2,500.
The child paternal allele was detected in 3 of the 30 mothers whose newborn was heterozygous at the loci examined.
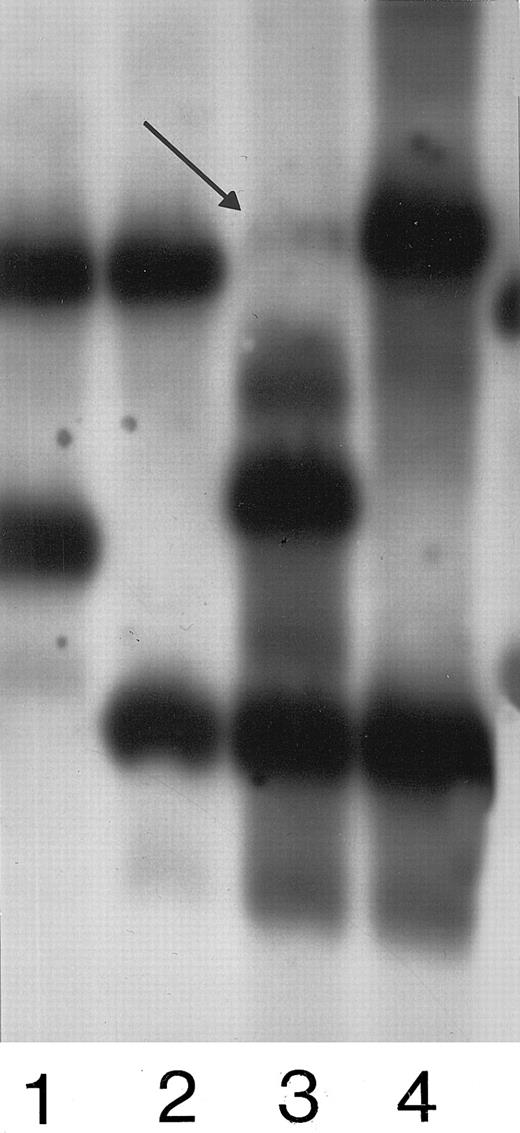
Figure 1 reports an example of ApoB minisatellite analysis.
Lane 1 shows a noncontaminated cord examined with ApoB. Lane 2 shows the respective mother. Lane 3 reports a contaminated cord. Lane 4 shows the mother. The arrow indicates the band due to maternal DNA contamination.
Lane 1 shows a noncontaminated cord examined with ApoB. Lane 2 shows the respective mother. Lane 3 reports a contaminated cord. Lane 4 shows the mother. The arrow indicates the band due to maternal DNA contamination.
The problem of contamination of CB units with maternal lymphocytes has been widely investigated. However, to our knowledge, the results available9-13 refer to numerically limited series and, to a certain extent, are discordant depending on the method used.
The method we used was able to detect maternal contamination from as few as 2 × 104 nucleated cells with a sensitivity of 0.04%, comparable to that reported by Sociè et al,12 without resorting to radioactivity.
Moreover, we were able to demonstrate an exchange of cells from child to mother through the placenta/uterus interface, supporting previously published studies that have shown the passage of fetal cells into the maternal circulation during pregnancy.10 14
However, we are aware that our series is limited, too; the combination of minisatellite amplification and chemiluminescence has never been used to this purpose and it proved to be highly sensitive.
ACKNOWLEDGMENT
This study was supported in part by a grant of Istituto Superiore di Sanità, Primo progetto di ricerca “Sangue 1995”-sottoprogetto No 3.3.1 “Standardizzazione dei metodi di raccolta, processazione e caratterizzazione delle cellule staminali da sangue placentare.”

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal