ONE OF THE MOST notable achievements of biomedical research in the first half of this century was the identification of red blood cell (RBC) antigens and the recognition of their importance to transfusion medicine and hemolytic disease of the newborn.1,2 These discoveries gave rise to an era of descriptive RBC serology during which a complex array of RBC membrane antigens were defined by their reactivity with specific alloantibodies.3,4 In recent years, RBC immunohematology has progressed from descriptive serology into an era of structural and functional analysis of blood group antigens, many of which are expressed in both erythroid and nonerythroid tissue.5-8 Here we will focus on the Duffy blood group antigen, a structure that has been of particular interest because it serves as a receptor on the RBC for the malarial parasite, Plasmodium vivax (P vivax ).8-10 We will attempt to highlight recent advances in our understanding of the molecular basis for the interaction of the P vivax malaria parasite with the Duffy blood group antigen and indicate how this information also contributed to the identification of an important gene family for cytoadherence proteins of P falciparum. In the context of normal physiology, the Duffy blood group antigen has been shown to be receptor for chemoattractant cytokines, or chemokines, and to be expressed by endothelial cells of postcapillary venules and by Purkinje cells of the cerebellum.11 12 We will attempt to review this important area of chemokine receptor research and discuss the potential relevance of the Duffy chemokine receptor to immunology and neurobiology.
Duffy Blood Group Serology
The Duffy blood group system first came to light in a report by Cutbush et al13 describing an alloantibody against an antigen denoted as Fya in a patient with hemophilia who had received multiple transfusions. The antithetical antigen, Fyb, was described 1 year later.14 Three phenotypes were identified in whites using anti-Fya and anti-Fyb antisera: Fy(a+b−), Fy(a−b+), and Fy(a+b+).15 These phenotypes are the products of codominant alleles comprising genotypes FY*A/FY*A, FY*B/FY*B, and FY*A/FY*B, respectively. Some whites express a weak or quantitatively reduced Fyb, the genetic basis of which remains uncertain.16
Most West Africans and 68% of African Americans do not express Fya or Fyb on their RBCs.17,18 The absence of Fya and Fyb on erythrocytes is designated the Duffy negative phenotype and denoted as Fy(a−b−). When Duffy-negative individuals of African descent develop anti-Duffy alloantibodies, they are almost always anti-Fya rather than anti-Fyb.19,20 The genotype designated FY/FY, which gives rise to the Fy(a−b−) erythroid phenotype, contains a coding sequence identical to that of FY*B.21 In individuals of the Fy(a−b−) phenotype, this sequence remains silent in erythroid cells but is transcribed and expressed on endothelial cells of postcapillary venules.22
Reactivity with anti-Fya and anti-Fyb alloantibodies is abolished after chymotrypsin or papain treatment of intact RBCs.23,24 Albrey et al25 described an anti-Duffy alloantibody, denoted anti-Fy3, that reacts with chymotrypsin-and papain-treated RBCs. Anti-Fy3 reacts with both Fy(a+b−) and Fy(a−b+) RBCs, but not Fy(a−b−) RBCs. Adsorption and elution experiments showed that anti-Fy3 reacts with a site common to both Fy(a+b−) and Fy(a−b+) RBCs. Although initially described in the serum of a rare Fy(a−b−) white woman (AZ) with a history of pregnancy and blood transfusions, anti-Fy3 can also occur in Duffy-negative individuals of African descent, often in conjunction with anti-Fya.18 Other Duffy-related epitopes have been defined by rare antisera, anti-Fy4 and anti-Fy5.26,27 Anti-Fy4 reacts only with Fy(a−b−) RBCs from individuals of African descent.26 Anti-Fy5 reacts with all human RBCs except those that are Fy(a−b−) and Rh null or express a variant of the e antigen of the Rh system.27 The rarity of these antisera has impeded progress in characterizing their epitopes.
THE DUFFY ANTIGEN IS A GLYCOPROTEIN
Moore et al28 showed that when surface-radioiodinated Fy(a+b−) RBCs were incubated with anti-Fya before solubilization in detergent, a protein of 35 to 43 kD was specifically immunoprecipitated. Coprecipitation of a radiolabeled protein other than the Duffy antigen was not formally excluded. More conclusive Western blotting experiments confirmed that a 35- to 43-kD protein carries the Fya antigenic determinant.29 Desialylation of RBC membranes with Vibrio cholera neuraminidase resulted in an altered electrophoretic mobility of the 35- to 43-kD Fya protein, suggesting that the Duffy antigen is a glycoprotein.30 Treatment of RBC membranes with N-glycanase caused a shift in the apparent molecular weight of the Duffy protein to around 30 kD.30,31 Although treatment of intact RBCs with trypsin did not proteolytically cleave the Duffy antigen, treatment of partially purified Duffy antigen with trypsin resulted in a 28-kD fragment.32 After incremental digestion of this 28-kD Duffy tryptic peptide with N-glycanase, Western blotting resolved multiple species, indicating that this glycoprotein has two or three asparagine-linked oligosaccharide chains.33 Parallel digestion with O-glycanase had no effect on the electrophoretic mobility of the glycoprotein, signifying the absence of oligosaccharide chains linked to serine and threonine residues.
MONOCLONAL ANTIBODIES (MoAbs) TO THE DUFFY ANTIGEN
In 1987, Nichols et al34 reported on the first MoAb (NYBC-BG6) against the Duffy antigen. This MoAb defined a new epitope, denoted Fy6, which is common to Fya and Fyb, but absent from Fy(a−b−) erythrocytes. Treatment of intact RBCs with chymotrypsin destroyed the Fy6 epitope as well as Fya and Fyb.35 An anti-Fy6 MoAb (i3A) similar to NYBC-BG6 was produced by Riwom et al36 and a MoAb (CBC-512) with a specificity similar to that of anti-Fy3 (reactivity not destroyed by treatment of RBCs with chymotrypsin or papain) was produced by Dr M. Uchikawa at the Japanese Red Cross (see Acknowledgment) (Table 1).
Available Human Polyclonal and Murine Monoclonal (MoAb) Anti-Duffy Antibodies
| Antibody . | Reactive RBCs . | Nonreactive RBCs . | Sensitive (S) or Resistant (R) . |
|---|---|---|---|
| . | . | . | to Chymotrypsin . |
| Anti-Fya (human) | Fy(a+b−), Fy(a+b+) | Fy(a−b+), Fy(a−b−) | S |
| Anti-Fyb (human) | Fy(a−b+), Fy(a+b+) | Fy(a+b−), Fy(a−b−) | S |
| Anti-Fy3 (human; MoAb) | Fy(a+b−), Fy(a−b+) Fy(a+b+) | Fy(a−b−) | R |
| Anti-Fy6 (MoAb) | Fy(a+b−), Fy(a−b+) Fy(a+b+) | Fy(a−b−) | S |
| Antibody . | Reactive RBCs . | Nonreactive RBCs . | Sensitive (S) or Resistant (R) . |
|---|---|---|---|
| . | . | . | to Chymotrypsin . |
| Anti-Fya (human) | Fy(a+b−), Fy(a+b+) | Fy(a−b+), Fy(a−b−) | S |
| Anti-Fyb (human) | Fy(a−b+), Fy(a+b+) | Fy(a+b−), Fy(a−b−) | S |
| Anti-Fy3 (human; MoAb) | Fy(a+b−), Fy(a−b+) Fy(a+b+) | Fy(a−b−) | R |
| Anti-Fy6 (MoAb) | Fy(a+b−), Fy(a−b+) Fy(a+b+) | Fy(a−b−) | S |
DUFFY IS INVOLVED IN P VIVAX PATHOPHYSIOLOGY
As a background to discussing the Duffy antigen in relation to malaria, a brief overview of the malarial life cycle will be presented. Malaria is transmitted by female anophiline mosquitos. When an infected mosquito takes a blood meal, sporozoites enter the blood and invade hepatocytes where they divide into thousands of merozoites. This part of the life cycle produces no symptoms in the host. Merozoites emerge from the liver and enter the the blood where they invade erythrocytes. This stage of the life cycle is associated with the clinical illness. Inside erythrocytes, merozoites develop sequentially into ring forms, trophozoites, and schizonts; mature schizonts give rise to many new merozoites. Infected erythrocytes ultimately lyse (“rupture”) and emerging merozoites invade other erythrocytes. Some ring forms develop into gametocytes instead of schizonts. Gametocytes are responsible for the transmission of malaria back to the mosquito when the mosquito takes a blood meal.
There are four species of human malaria: P falciparum,P vivax,P ovale, and P malariae. Of these, P falciparum and P vivax are the most prevalent. P falciparum causes the most mortality; P vivax is second to P falcipaurm as a cause of malaria-related morbidity. P knowlesi, a primate malaria related to P vivax and capable of invading human RBCs, has been used as a laboratory model to investigate how malaria parasites invade RBCs.
In 1975, Miller et al reported that human erythrocytes of the Fy(a−b−), or Duffy-negative phenotype, were resistant to invasion by merozoites (the RBC invasive form) of P knowlesi.37 38 Human erythrocytes of all other phenotypes were invaded. Furthermore, anti-Fya and anti-Fyb specifically inhibited invasion of P knowlesi merozoites into human RBCs of the Fy(a+b−) and Fy(a−b+) phenotype, respectively.37
Cytochalasin B–treated merozoites were used to study attachment independent of invasion.39 Cytochalasin B–treated merozoites of P knowlesi attached equally well to both Duffy-positive and Duffy-negative erythrocytes. Ultrastructural analysis of Duffy positive erythrocytes with adherent merozoites showed an electron dense structure beneath the RBC lipid bilayer in the area of apposition between RBC and merozoite.39 Previous studies had demonstrated that this structure, termed a tight junction, is of crucial importance for parasite invasion.40 The tight junction was not observed with merozoites adherent to Duffy-negative erythrocytes.
The finding that P knowlesi requires the Duffy antigen to form a tight junction and to invade human erythrocytes suggested an explanation for two observations related to the human malaria, P vivax. First, P vivax, which is prevalent in most tropical and subtropical areas of the world, is specifically absent from West Africa, a geographic area where greater than 95% of individuals are Fy(a−b−). Second, during treatment of neurosyphilitic patients with therapeutically induced malaria, many African Americans were noted to be innately resistant to P vivax.41 Miller et al42 postulated that the innate resistance of some African Americans to P vivax malaria might be related to the Duffy-negative phenotype. A number of studies confirmed this hypothesis and demonstrated a correlation between the Fy(a−b−) phenotype and resistance to P vivax infection.42-44 It was subsequently shown that Fy(a−b−) erythrocytes cannot be invaded by P vivax in vitro and that the anti-Fy6 MoAb, NYBC-BG6, can block invasion of Duffy-positive erythrocytes by P vivax.34 35
One of the distinguishing features of P vivax is its preference for reticulocytes.45 Because the Duffy antigen is expressed on mature erythrocytes as well as on reticulocytes, it has been postulated that in addition to the Duffy glycoprotein, P vivax parasites require a second RBC receptor for invasion that is specific for reticulocytes. Although the reticulocyte-specific RBC receptor for invasion by P vivax has not yet been identified, a P vivax ligand that binds specifically to reticulocytes has been identified and cloned.46
DUFFY BINDING LIGANDS OF PKNOWLESI AND PVIVAX
Haynes et al47 identified a parasite protein (Duffy binding ligand) from P knowlesi that binds specifically to Duffy-positive erythrocytes but not to Duffy-negative erythrocytes. The 135-kD Duffy binding ligand of P knowlesi also binds to rhesus RBCs, which serologically react as Fy(a−b+) and which express an Fyb-related antigen that is highly homologous, but not identical to, human Fyb.21,47 A similar Duffy binding ligand of 140 kD was isolated by Wertheimer and Barnwell48 from P vivax. Unlike the 135-kD Duffy binding protein of P knowlesi, the 140-kD Duffy binding protein of P vivax does not bind to rhesus RBCs, a cell type that cannot be invaded by P vivax.48 Recent molecular analyses of the Duffy binding ligands of P knowlesi,P vivax, and the erythrocyte binding ligand of P falciparum (which does not bind to Duffy but to a sialic acid–dependent domain on glycophorin A) indicate regions of homology among these erythrocyte binding proteins.49 50
Antibodies prepared against the 135-kD Duffy binding ligand of P knowlesi were used to screen expression complementary DNA (cDNA) libraries prepared from P knowlesi asexual erythrocyte-stage mRNA and cDNA encoding the 135-kD Duffy binding protein was cloned.51 The highly related 140-kD Duffy binding protein of P vivax was identified from cDNA clones obtained by cross-hybridization with the P knowlesi cDNA probe.52 Immunohistochemical studies localized the Duffy binding protein of P knowlesi to the micronemes, specialized organelles at the apical end of the parasite (merozoite) that appear to function in the process of invasion. This further supported the conclusion that Duffy binding proteins are indeed parasite ligands that bind to the Duffy receptor during invasion.51 How these proteins are translocated from micronemes to the apical surface of the merozoite to interact with the Duffy receptor is unknown.
The extracellular regions of the 140-kD Duffy binding protein of P vivax and the 135-kD Duffy binding protein of P knowlesi were classified into six domains based on amino acid sequence similarities.51,53 COS 7 cells were transfected with constructs encoding region I, region II, regions III-V, and region VI, each designed to yield cell-surface expression.53 Cells expressing the cysteine-rich region II specifically formed rosettes with Duffy-positive, but not Duffynegative, erythrocytes, indicating that this domain is responsible for interactions with erythrocytes mediated by the Duffy glycoprotein.53
Although the 140-kD Duffy binding ligand of P vivax did not bind to rhesus RBCs in the rosetting assay, binding did occur if rhesus RBCs were first treated with N-glycanase.54 Synthetic peptides consisting of the first 35 amino acids from the N-terminus of the human Duffy protein and the first 34 amino acids from the N-terminus of the rhesus Duffy homolog both blocked rosetting of Duffy-positive human erythrocytes with COS cells expressing the 140-kD Duffy binding protein of P vivax.54 These data support the conclusion that the Duffy homolog on rhesus RBCs contains the peptide binding site for the P vivax Duffy binding protein. However, it appears that on the intact rhesus RBCs, this site is altered or obscured by the presence of one or more N-linked sugar side chains. Thus, it appears that differences in glycosylation may contribute to species specificity of malaria parasite-host cell interactions.
DUFFY BINDING LIGAND MOTIFS IN THE VAR GENES OF PFALCIPARUM
Identification of the Duffy binding proteins of P knowlesi and P vivax provided a clue to the identification of another important malarial gene family, termed the var genes.55-58 Howard et al59-61 had shown that malaria parasites, including P falciparum, insert parasite-derived proteins into the membranes of infected RBCs. Biologic data indicated that one or more of these proteins function in cytoadherence of P falciparum–infected RBCs to endothelial cells.62,63 Cytoadherence of P falciparum–infected RBCs to endothelial cells of postcapillary venules accounts for the sequestration of mature parasite-infected RBCs in the deep vasculature and their consequent absence on diagnostic blood films.64,65 Ultrastructural studies of sequestered parasites showed that the points of contact between infected RBCs and endothelial cells consist of electron dense knobs on the RBC membranes.66,67 Sequestration allows parasites to grow within the vasculature of the host without circulating through the spleen, an organ known to have anti-malarial properties.67,68 Cytoadherence of infected RBCs to endothelial cells of cerebral vasculature also contributes to the pathogenesis of cerebral malaria.69 David et al70 demonstrated that treatment of P falciparum–infected monkeys with hyperimmune serum reversed sequestration in a strain-specific fashion and led to the appearance of crisis forms (morphologically deteriorating parasites within erythrocytes). Serologic and immunochemical evidence indicated that the endothelial binding proteins on the surface of parasite-infected RBCs undergo antigenic variation, a phenomenon of obvious importance to the parasite as it affords a mechanism of circumventing the effects of specific antibodies that might otherwise block cytoadherence.71-75 Understanding the molecular basis for endothelial cytoadherence and antigenic variation was thwarted for years by difficulties purifying sufficient quantities of the malarial endothelial cytoadherence protein for microsequencing. Unexpectedly, analysis of Duffy binding ligands provided a clue to the indentification of genes encoding variant endothelial cytoadherence proteins of P falciparum.55 56
While sequencing DNA from regions of parasite chromosome 7, Peterson et al55 and Su et al56 identified several gene sequences that contain domains similar to region II of the erythrocyte binding proteins of P knowlesi,P vivax, and P falciparum. This family of related sequences possessed properties predicted for genes encoding cytoadherence proteins: they were polymorphic, they contained large open reading frames consistent with the size of putative cytoadherence proteins, and their predicted protein products included multiple domains with regions homologous to Duffy binding proteins, which had already been shown to have cytoadhesive properties. Taking advantage of homologies within these Duffy binding-like domains, Smith et al57 used degenerate oligonucleotide primers to study messenger RNA from these genes (designated variant or var genes) expressed in cloned parasites with serologically distinct surface antigens and cytoadherence properties. They showed that the expression of distinct cytoadhesive properties and antigenic reactivity at the surface of infected erythrocytes correlated with expression of distinct var genes.57 Baruch et al58 cloned cDNA from an expression library for the putative cytoadherence protein, PfEMP1 (P falciparum erythrocyte membrane protein 1), and demonstrated by immunochemical methods that peptides derived from the cDNA had antigenic properties predicted for a cytoadherence protein. These investigators, together with Su et al, showed that cDNA for PfEMP1 is a member of the var gene family.57,58 The studies by Su et al,56 Smith et al,57 and Baruch et al58 were published simultaneously and confirmed that var genes encode related, but antigenically variant, cytoadherence proteins.
Malarial Receptor/Adhesion Proteins: Potential Targets of Receptor/Adhesion Blocking Immunity
| Parasite . | Receptor/Adhesion Protein . | Location . | Host Counter Receptor . |
|---|---|---|---|
| P knowlesi | 135-kD Duffy binding protein (for invasion) | Merozoite* | Duffy glycoprotein (DARC) on RBCs |
| P vivax | 140-kD Duffy binding protein (for invasion) | Merozoites | Duffy glycoprotein (DARC) on RBCs |
| P falciparum† | 170-kD Erythrocyte binding protein (for invasion) | Merozoites | Sialic Acid-glycophorin A on RBCs |
| P falciparum | var gene products (for sequestration) | Membrane of infected RBCs | CD36, ICAM, VCAM, E-selectin (? thrombospondin) on endothelial cells |
| Parasite . | Receptor/Adhesion Protein . | Location . | Host Counter Receptor . |
|---|---|---|---|
| P knowlesi | 135-kD Duffy binding protein (for invasion) | Merozoite* | Duffy glycoprotein (DARC) on RBCs |
| P vivax | 140-kD Duffy binding protein (for invasion) | Merozoites | Duffy glycoprotein (DARC) on RBCs |
| P falciparum† | 170-kD Erythrocyte binding protein (for invasion) | Merozoites | Sialic Acid-glycophorin A on RBCs |
| P falciparum | var gene products (for sequestration) | Membrane of infected RBCs | CD36, ICAM, VCAM, E-selectin (? thrombospondin) on endothelial cells |
135-kD Duffy binding protein has been localized to apical organelles (micronemes) of the merozoite.
The 170-kD erythrocyte binding protein and the var gene products of P falciparum contain Duffy binding-like domains.
Identification of var genes of P falciparum represents a major advance in malaria research because it provides the molecular basis for understanding sequestration, antigenic variation, and the chronicity of malaria. There is evidence that each parasite of P falciparum contains a large repertoire of var genes (50 to 150 copies) scattered throughout the malarial genome.57 These genes, which encode highly homologous but antigenically distinct cytoadherence proteins, account for 2% to 6% of the haploid parasite genome.57 Despite antigenic differences among these variant cytoadherence proteins of P falciparum, their capacity to bind to endothelial cells is conserved.56 The molecular basis for this conservation of function in the face of antigenic variation remains to be elucidated, although it is clear that a number of endothelial receptors, including CD36, ICAM, VCAM, E-selectin, and thrombospondin may be involved in cytoadherence.76-78
THE IMPORTANCE OF MALARIAL CYTOADHERENCE TO VACCINE RESEARCH
Cytoadherence of all species of malarial merozoites to host RBCs, followed by invasion, is crucial for the life cycle of these obligate intracellular parasites. Cytoadherence of P falciparum–infected RBCs to endothelial cells of postcapillary venules appears crucial to the survival of this particular parasite. The parasite proteins discussed above are part of a newly described superfamily of malarial cytoadherence molecules (Table 2). If these molecules could be used as immunogens to induce antibodies that block cytoadherence, protection against malaria might be achieved (Fig 1). Even if the protection were partial, it would potentially be beneficial because many of the pathologic sequelae of malarial infections in the partially immune host are related to the level of parasitemia.79 Because the endothelial cytoadherence molecule of P falciparum is highly variant, it will present a major challenge to development of a malaria vaccine. The parasite ligands that bind to RBC receptors appear much less variant, although there is evidence that P falciparum can invade by more than one pathway and can switch between at least two alternative pathways for invasion.80 Although there is some variation within the adhesion domain of the P vivax Duffy binding protein, this parasite appears to be absolutely dependent on the Duffy pathway for invasion.81P vivax provides a model for testing the efficacy of an erythrocyte binding protein–based vaccine, especially now that inhibition of ligand-receptor interaction by immune sera can be tested in vitro using the transfected COS cell-RBC rosetting assay established by Chitnis et al.53 54
(A) Two receptors for apical attachment of P vivax merozoites to RBCs have been cloned: the Duffy binding protein and the reticulocyte binding protein.46,52 One strategy for vaccine development is to use these proteins as immunogens to induce antibodies that might block invasion of the parasite into the RBC. (B) Variant cytoadherence proteins of P falciparum have also been cloned.55-57 Antibodies against this protein can block cytoadherence of P falciparum–infected RBCs to endothelial cells, forcing the parasite to circulate through the spleen, where immunologic mechanisms (which have not been clearly defined) are brought into play against P falciparum parasites. The cytoadherence antigen is variant and antigenic variation is used by the parasite to escape antibody mediated blockade of cytoadherence. Another strategy for vaccine development is to identify domains that are shared among different variants of the cytoadherece protein of P falciparum; antibodies against shared domains could block cytoadherence in a variant-independent fashion.
(A) Two receptors for apical attachment of P vivax merozoites to RBCs have been cloned: the Duffy binding protein and the reticulocyte binding protein.46,52 One strategy for vaccine development is to use these proteins as immunogens to induce antibodies that might block invasion of the parasite into the RBC. (B) Variant cytoadherence proteins of P falciparum have also been cloned.55-57 Antibodies against this protein can block cytoadherence of P falciparum–infected RBCs to endothelial cells, forcing the parasite to circulate through the spleen, where immunologic mechanisms (which have not been clearly defined) are brought into play against P falciparum parasites. The cytoadherence antigen is variant and antigenic variation is used by the parasite to escape antibody mediated blockade of cytoadherence. Another strategy for vaccine development is to identify domains that are shared among different variants of the cytoadherece protein of P falciparum; antibodies against shared domains could block cytoadherence in a variant-independent fashion.
THE DUFFY ANTIGEN IS A NOVEL RECEPTOR FOR CHEMOATTRACTANT CYTOKINES
Thus far we have discussed the function of the Duffy antigen in the pathogenesis of malaria. We have attempted to describe how knowledge of the Duffy antigen and its relationship to P knowlesi and P vivax malaria led to identification of malarial Duffy binding ligands, and this in turn provided clues to the identification of endothelial binding ligands of P falciparum. But clearly the Duffy glycoprotein exists for purposes other than malarial invasion of RBCs. Unexpectedly, research on chemoattractant cytokines, abbreviated chemokines, and their receptors converged with investigation on the Duffy blood group antigen, providing insight into a potential normal physiologic role for this allelic glycoprotein.10 11 To put this research into proper perspective, it is necessary to briefly review the biologic activities of chemokines.
Chemokines are members of a superfamily of small, secreted proteins (≈8 to 13 kD), numbering over 20, that recruit leukocytes to sites of inflammation.82 This superfamily has two major branches, encoded in separate gene clusters, that preferentially promote acute and chronic inflammatory processes.83-85 In addition to the functional differences between the two families, they are also distinguished by structural characteristics and at the level of genomic organization.83-85 Chemokines of the C-X-C family, in which the two amino proximal cysteine residues are separated by one amino acid, function primarily as chemoattractants for neutrophils, whereas chemokines of the C-C family, in which these cysteines are juxtaposed, function primarily as chemoattractants for lymphocytes and monocytes. A newly discovered chemokine, lymphotactin, has a single cysteine, or C-motif rather than a C-C or C-X-C motif.86
The effects of chemokines are mediated through high-affinity receptors that have seven hydrophobic membrane-spanning helices and signal through coupling with G-proteins.87-92 In general, each chemokine has a specific receptor to which it binds with high affinity and another receptor to which it binds with lower affinity and with which it shares binding with another chemokine of the same family. In 1991, Darbonne et al93 discovered a novel chemokine receptor on RBCs. Analysis of the repertoire of ligands bound by this novel receptor showed that it binds selected members of both the C-X-C and C-C families with high affinity (Kd ≈ 5 nM).94,95 These include interleukin-8 (IL-8) and melanoma growth stimulatory activity (MGSA) from the C-X-C family and RANTES (regulated upon activation, normally T-cell expressed) and monocyte chemotactic peptide (MCP-1) from the C-C family.94,95 The RBC chemokine receptor does not bind the C chemokine, lymphotactin.96 The multispecific erythrocyte chemokine binding activity is preserved in other mammalian species, including mice, and in avian species.11,21 96
Further studies on the multispecific chemokine receptor on human RBCs led to the observation that approximately two thirds of African Americans lack this receptor activity, a frequency that was intriguingly similar to that of the Duffy-negative phenotype. Subsequent investigation led to the conclusion that the RBC chemokine receptor and the Duffy blood group antigen are identical.97 There was an absolute correlation between chemokine binding activity and Duffy blood group antigen expression. Anti-Fy6 specifically blocked binding of chemokines to Duffy positive RBCs (Fig 2). Both the RBC chemokine receptor and the Duffy blood group antigen were destroyed by chymotrypsin treatment, but not trypsin treatment, of intact RBCs. Cross-linking experiments with 125I-labeled MGSA indicated that the RBC chemokine receptor is a 35- 43-kD protein with a similar appearance on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as that of the Duffy antigen when detected by immunoblotting with anti-Fy6. MGSA, IL-8, and a mutant MGSA, designated MGSA-E6A, which evinces high-affinity binding only to the RBC chemokine receptor, were shown to block invasion of human RBCs by P knowlesi, a parasite known to use the Duffy antigen for invasion.97,98 MGSA was also shown to block binding of the 135-kD Duffy binding protein of P knowlesi to Duffy-positive human RBCs.97 The conclusion drawn from this evidence that the human RBC multispecific chemokine receptor and the Duffy blood group antigen are identical was subsequently confirmed by transfection experiments. K562 and 293 cells (both of which normally lack the Duffy antigen and are devoid of a multispecific chemokine receptor) were transfected with cloned Duffy cDNA and consequently displayed concomitant Duffy antigenic and multispecific chemokine receptor activity on their surfaces.99 100
Inhibition of chemokines binding to the DARC by the anti-Duffy MoAb, anti-Fy6. (A) Inhibition of specific 125I-labeled IL-8 binding to Duffy-positive RBCs by increasing concentrations of anti-Fy6. The closed circles represent binding of 125I-labeled IL-8 to cells expressing type A IL-8 receptor. Anti-Fy6 specifically inhibits binding of IL-8 to DARC in a dose-dependent fashion. (B) Inhibition of specific binding of radiolabeled IL-8, MGSA, MCP-1, and RANTES to Duffy-positive RBCs by anti-Fy6. Anti-Fy6 inhibits the binding of both C-X-C chemokines (IL-8, MGSA) and C-C chemokines (MCP-1, RANTES) to DARC. (Reprinted with permission from Science, Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, Miller LH: A receptor for the malarial parasite Plasmodium vivax: The erythrocyte chemokine receptor. 261:1182, 1993. Copyright 1993 American Association for the Advancement of Science.)97
Inhibition of chemokines binding to the DARC by the anti-Duffy MoAb, anti-Fy6. (A) Inhibition of specific 125I-labeled IL-8 binding to Duffy-positive RBCs by increasing concentrations of anti-Fy6. The closed circles represent binding of 125I-labeled IL-8 to cells expressing type A IL-8 receptor. Anti-Fy6 specifically inhibits binding of IL-8 to DARC in a dose-dependent fashion. (B) Inhibition of specific binding of radiolabeled IL-8, MGSA, MCP-1, and RANTES to Duffy-positive RBCs by anti-Fy6. Anti-Fy6 inhibits the binding of both C-X-C chemokines (IL-8, MGSA) and C-C chemokines (MCP-1, RANTES) to DARC. (Reprinted with permission from Science, Horuk R, Chitnis CE, Darbonne WC, Colby TJ, Rybicki A, Hadley TJ, Miller LH: A receptor for the malarial parasite Plasmodium vivax: The erythrocyte chemokine receptor. 261:1182, 1993. Copyright 1993 American Association for the Advancement of Science.)97
DUFFY ANTIGEN RECEPTOR FOR CHEMOKINES (DARC) IS EXPRESSED BY ENDOTHELIAL CELLS OF POSTCAPILLARY VENULES
The physiologic significance of the novel multispecific RBC chemokine receptor, designated the Duffy antigen receptor for chemokines (DARC), initially appeared questionable because no pathologic or inflammatory consequences have been associated with the Duffy-negative phenotype. However, immunohistochemical staining of human tissues with anti-Fy6 provided additional insight into a potential physiologic function for DARC.22,101 Specific and intense staining with anti-Fy6 was observed with endothelial cells lining postcapillary venules throughout the body. Staining was not observed on endothelial cells lining capillaries or larger vessels, including venules, veins, arterioles, and arteries.101a Littoral cells, specialized endothelial cells that line the sinusoids in the red pulp of the spleen, were also strongly positive (Fig 3) as were the endothelial cells that line the bone marrow sinusoids and choroid plexus. Parallel chemokine binding experiments confirmed the presence of a high-affinity receptor having a promiscuous chemokine binding repertoire in membrane fractions from kidney and spleen.22,100 Endothelial cells lining the hepatic sinusoids lacked immunoreactivity. Tissue from individuals of the Fy(a−b−) erythroid phenotype were also examined and found to react with anti-Fy6 in a fashion identical to tissue from Duffy-positive individuals.22 Interestingly, endothelial cells of larger vessels were observed to react with anti-Fy6 under conditions of inflammation (eg, temporal arteritis, thrombophlebitis, omphalitis), suggesting upregulation of DARC under these conditions.101a
Immunohistochemistry of spleen using the anti-Duffy MoAb, anti-Fy6. Analysis was done on both freshly obtained and archival specimens of spleen. Anti-Fy6 reacts with specialized endothelial cells (littoral cells) lining the sinusoids in the red pulp. Endothelial cells lining arterioles, venules, arteries, and veins did not stain with anti-Fy6. Similar staining was observed with endothelial cells of postcapillary venules in every organ examined thus far. Immunohistochemical staining of endothelial cells in tissue obtained from Duffy-negative individuals (by RBC typing) was identical to that observed in tissue obtained from Duffy-positive individuals. In contrast to the sinusoids of the spleen, endothelial cells lining hepatic sinusoids did not stain.
Immunohistochemistry of spleen using the anti-Duffy MoAb, anti-Fy6. Analysis was done on both freshly obtained and archival specimens of spleen. Anti-Fy6 reacts with specialized endothelial cells (littoral cells) lining the sinusoids in the red pulp. Endothelial cells lining arterioles, venules, arteries, and veins did not stain with anti-Fy6. Similar staining was observed with endothelial cells of postcapillary venules in every organ examined thus far. Immunohistochemical staining of endothelial cells in tissue obtained from Duffy-negative individuals (by RBC typing) was identical to that observed in tissue obtained from Duffy-positive individuals. In contrast to the sinusoids of the spleen, endothelial cells lining hepatic sinusoids did not stain.
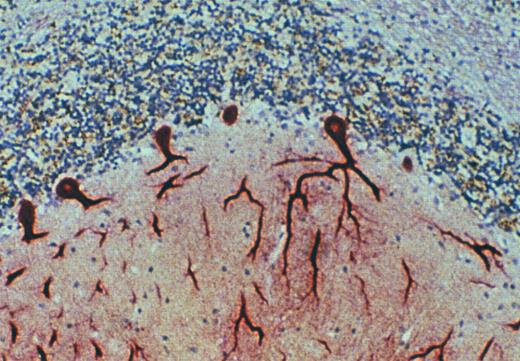
Purkinje cells of the cerebellum express the DARC. Immunohistochemical staining of archival specimens of human cerebellum with the anti-DARC MoAb, anti-Fy6, showed high-level expression of DARC by Purkinje neurons. This staining was inhibited by a recombinant fusion protein in which the amino terminal extracellular domain of DARC was expressed in continuous translational frame with glutathione-S-transferase. Cross-linking experiments with 125I-labeled MGSA and immunoblots with anti-Fy6 showed that MGSA and anti-Fy6 react with a protein component of cerebellar membranes with the same size and appearance on SDS-PAGE as RBC and endothelial cell DARC (data not shown).
Purkinje cells of the cerebellum express the DARC. Immunohistochemical staining of archival specimens of human cerebellum with the anti-DARC MoAb, anti-Fy6, showed high-level expression of DARC by Purkinje neurons. This staining was inhibited by a recombinant fusion protein in which the amino terminal extracellular domain of DARC was expressed in continuous translational frame with glutathione-S-transferase. Cross-linking experiments with 125I-labeled MGSA and immunoblots with anti-Fy6 showed that MGSA and anti-Fy6 react with a protein component of cerebellar membranes with the same size and appearance on SDS-PAGE as RBC and endothelial cell DARC (data not shown).
As noted previously, DARC-like molecules are expressed on RBCs of rodents and other mammalian and avian species.11,21,96 Because many model systems for the actions of chemokines on leukocyte trafficking, angiogenesis, and microvascular leak syndromes have been developed in rodents, experiments were performed to determine whether DARC is expressed by endothelial cells of postcapillary venules in rats. Because immunologic reagents for the rodent homolog of DARC are not yet available, intravital microscopy was performed on rats infused with fluorescent microspheres covalently coated with IL-8 or MCP-1 into the cremaster vessels.101a Monitoring of fluorescent microsphere circulation revealed attachment to endothelial cells lining small venules, but not to larger vessels. This binding of IL-8–coated microspheres was inhibited by previous infusion of IL-8, as well as MCP-1. Control microspheres did not adhere to endothelial cells. These experiments provide indirect functional data that the rodent homolog of DARC is expressed by subsets of endothelium according to a program that mimics that observed in humans.
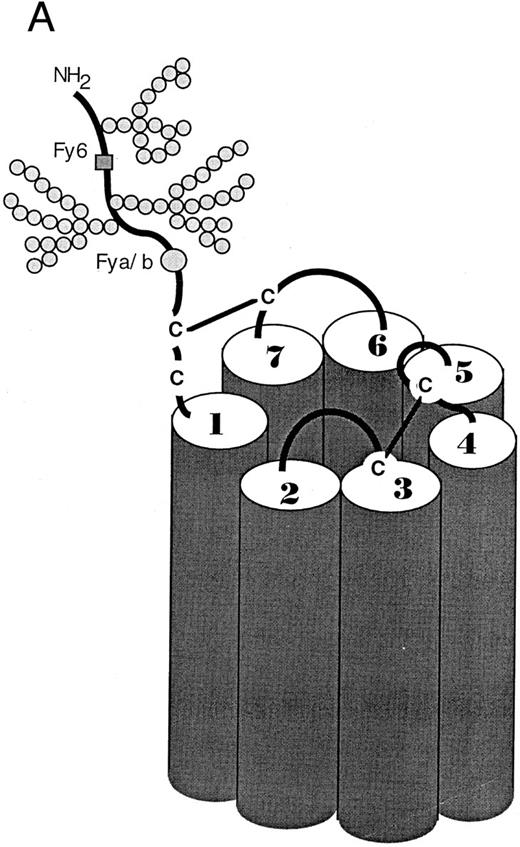
Proposed seven transmembrane spanning topology of DARC. (A) Molecular modeling predicts that DARC has topologic features similar to other members of the chemokine receptor family. (Modified and reprinted with permission.113 ) The amino terminal extracellular domain contains the binding site for anti-Fy6 MoAbs and the polymorphism resulting in the Fya and Fyb blood groups. This domain has also been found to contain sequences necessary for multi-specific chemokine binding. Sequences required for binding a MoAb with anti-Fy3 specificity have been tentatively localized to the third predicted extracellular loop. (B) The deduced amino acid sequence of DARC; the first seven N-terminal amino acids shown are those predicted from the exon identified by Iwamoto111 and are thought to be present in the major DARC transcript; the N-terminal amino acids shown in parentheses and the rest of the amino acid sequence are derived from the single DARC exon as originally cloned by Chaudhuri et al.105 (Modified and reprinted with permission from Horuk R: Molecular properties of the chemokine receptor family. Trends in Pharmacological Sciences, vol 15, p 159, 1994.88 ).
Proposed seven transmembrane spanning topology of DARC. (A) Molecular modeling predicts that DARC has topologic features similar to other members of the chemokine receptor family. (Modified and reprinted with permission.113 ) The amino terminal extracellular domain contains the binding site for anti-Fy6 MoAbs and the polymorphism resulting in the Fya and Fyb blood groups. This domain has also been found to contain sequences necessary for multi-specific chemokine binding. Sequences required for binding a MoAb with anti-Fy3 specificity have been tentatively localized to the third predicted extracellular loop. (B) The deduced amino acid sequence of DARC; the first seven N-terminal amino acids shown are those predicted from the exon identified by Iwamoto111 and are thought to be present in the major DARC transcript; the N-terminal amino acids shown in parentheses and the rest of the amino acid sequence are derived from the single DARC exon as originally cloned by Chaudhuri et al.105 (Modified and reprinted with permission from Horuk R: Molecular properties of the chemokine receptor family. Trends in Pharmacological Sciences, vol 15, p 159, 1994.88 ).
It is of note that the subset of endothelial cells that express DARC is similar to the anatomic site of leukocyte trafficking and are targets for both adhesion and diapedesis.102-104 In addition, endothelial cells in this anatomic distribution are targets for vascular leak syndromes induced by pathologic actions of IL-8. Thus, in addition to expression on erythrocytes, which is not essential, DARC is expressed by endothelial cells at a dynamic interphase of leukocyte trafficking, a process that is highly regulated by the actions of chemokines that bind to DARC. Whether or not DARC actually plays a role in leukocyte trafficking remains to be determined.
THE MOLECULAR BIOLOGY OF DARC
Complementary DNA (cDNA) encoding DARC was molecularly cloned by Chaudhuri et al105 at the New York Blood Center in 1993. Sequences of various peptides from the Duffy glycoprotein, which were purified by immunoaffinity chromatography with Fy6 MoAbs, were used to design oligonucleotide primers that were employed in DNA amplification reactions.105,106 A 72-bp probe generated by reverse transcription followed by polymerase chain reaction was used to screen a cDNA library prepared from bone marrow cells of a Duffy-positive individual. Nucleotide sequence analysis of overlapping cDNA clones revealed an open reading frame (ORF ) of 1,014 bp, which predicted a protein product of 338 amino acids, with a theoretical molecular weight of 35 kD. In situ hybridization confirmed previous linkage analysis that the Duffy locus is on chromosome 1q22 → q23.107,108 The Duffy cDNA was noted to share significant homology with the human and rabbit type B IL-8 receptor, which also binds MGSA at high affinity.105 Surprisingly, DARC also shares a similar level of homology to the multispecific receptor for endothelins, vaso-active proteins with 21 amino acid residues and two internal disulfide bonds, which are also expressed by endothelial cells and have a profound effect on vascular biology. Whereas the similarity between DARC and the IL-8RB is more prominent in the amino-terminal and carboxyl-terminal regions, its similarity to the multispecific endothelin receptor (type B) is evident in the loops (both extracellular and cytoplasmic) and transmembrane-spanning helices. Unlike virtually all other heptahelical receptors, which are almost universally linked to G-proteins, DARC lacks a highly conserved DRY motif in the second cytoplasmic loop.
Schematic representation of the DARC genomic locus. DARC is encoded by an ORF contained in two exons separated by one intron. Primer extension and 5′RACE analysis of both human DARC and mouse DARC homolog (Wang Z.X., Lu A.H., Peiper S.C., unpublished observations, September 1995) reveal the presence of an upstream exon containing a methionine translation initiation codon and codons for six additional amino acid residues. An internal initiation codon that may be used in the human gene is not conserved in the mouse gene. The polymorphism responsible for the Fya and Fyb phenotypes is encoded at codon 44.
Schematic representation of the DARC genomic locus. DARC is encoded by an ORF contained in two exons separated by one intron. Primer extension and 5′RACE analysis of both human DARC and mouse DARC homolog (Wang Z.X., Lu A.H., Peiper S.C., unpublished observations, September 1995) reveal the presence of an upstream exon containing a methionine translation initiation codon and codons for six additional amino acid residues. An internal initiation codon that may be used in the human gene is not conserved in the mouse gene. The polymorphism responsible for the Fya and Fyb phenotypes is encoded at codon 44.
Although molecular modeling approaches described in the original cloning report predicted the presence of nine hydrophobic helices that could serve as transmembrane spanning domains, subsequent molecular modeling analyses predicted the presence of seven hydrophobic α-helices, analogous to receptors for all of the other known chemoattractants.11,99 109 Although DARC undergoes cotranslational addition of asparagine-linked oligosaccharide chains and posttranslational remodeling of carbohydrates in the Golgi, the predicted primary structure lacks an N-terminal series of hydrophobic amino acid residues that could function as a signal peptide. This characteristic is common to all of the cloned chemokine receptors. It is presumed that these proteins are targeted to membrane-bound polyribosomes and the Golgi apparatus via the hydrophobic helices, which are predicted to serve as transmembrane anchors.
The presence of seven hydrophobic helices suggests a topology in which there are an amino-terminal extracellular domain (approximately 62 residues), three extracellular loops, three cytoplasmic loops, and a carboxly-terminal cytoplasmic tail (28 residues) (Fig 4). Alignment with the primary structures of other known chemokine receptors shows that DARC contains cysteine residues in each extracellular loop that line up with those present in the other receptors, as well as the presence of other highly conserved residues. The N-terminal extracellular domain contains at least two, and possibly three, sites for addition of oligosaccharide chains to asparagine residues. The N-terminal extracellular domain is rich in acidic amino acid residues, as might be expected for a domain involved in binding ligands with a basic isoelectric point (pI). As characteristic of other chemokines receptors, the cytoplasmic tail is composed of approximately 50% serine and threonine residues, which are presumed to be targets for phosphorylation, as has been observed for the type B IL-8 receptor.
The organization of the gene encoding DARC has not yet been completely elucidated. Using primer extension analysis, Chaudhuri et al105 found the RNA cap site to be approximately 175 bp upstream from the (presumed) translation initiation codon. Amplification of the ORF with primers from the 5′ and 3′ untranslated regions by polymerase chain reaction yielded DNA fragments of identical size when genomic DNA and cDNA reverse transcribed from mRNA served as the source of templates. Nucleotide sequence analysis confirmed that the ORF was contained within a single, uninterrupted exon. Constructs prepared from this ORF were capable of encoding a protein of the predicted size that bound anti-Duffy MoAbs and the predicted panel of chemokines at the appropriate affinity when expressed in K562 cells and human embryonic kidney cells.99,100 However, because the amino-terminal peptide sequence of the Duffy antigen has not been determined, it has not been confirmed that the translation initiation codon proposed by Chaudhuri et al is the predominant one used in vivo.105
Analysis by other groups provide evidence that for an mRNA cap site different from that initially reported. Using the method of rapid amplification of cDNA ends (5′RACE), Tournamille et al110 reported that the mRNA initiation site was 495 bp upstream of the translation initiation codon. Similar studies described by Iwamoto et al111 localized the cap site to −550 bp. The latter report also described a novel upstream exon that contained an alternative ATG codon. Direct comparison showed that this exon, which encodes a methionine and six additional amino acid residues, is preferentially utilized over the downstream cap site described by Chaudhuri et al. Analysis of the mouse gene encoding the homolog of DARC performed in our laboratory supports the findings of Iwamoto et al.111 The downstream methionine residue is not conserved in the mouse gene. 5′RACE of mRNA from mouse spleen confirmed the presence of an upstream exon encoding seven amino acids, led by a methionine residue. This exon, which is followed by a consensus splice donor sequence, is separated from the exon containing the majority of the ORF by an intron of approximately 450 bp. Together, these findings suggest that both human DARC and the mouse homolog are encoded by a gene composed of two exons and one intron (Fig 5).
The importance of DARC is evidenced by the conservation of this gene across species. We have cloned genes encoding DARC homologs from six nonhuman primates, cow, pig, rabbit, and mouse. There is over 95% conservation of primary structure between the DARC homologs in humans and great apes. There is some divergence in macaques. The nucleotide sequence of the rhesus homolog predicts six amino acid differences compared with humans and a deletion of codon 24 which encodes an aspartic acid residue in the amino-terminal extracellular domain.21 Rhesus RBCs react with anti-Fyb but not with anti-Fy6. There is divergence in the predicted amino acid sequence of the murine homolog to a level of approximately 65% identity with the human receptor protein.
PRESERVATION OF DARC EXPRESSION IN ENDOTHELIAL CELLS OF DUFFY-NEGATIVE INDIVIDUALS
Despite the conservation of DARC function and primary structure across species, the fact that a large population of individuals of African ancestry lacks expression of this receptor on their RBCs without any detectable adverse consequences represented a problem for those wishing to assign physiologic significance to DARC. Whereas Southern blotting experiments failed to show evidence of a gross rearrangement or deletion involving the DARC gene in DNA from Duffy-negative individuals, Northern blot analysis failed to detect mRNA transcripts encoding DARC in bone marrow (erythroid) cells.103 However, Northern blot analysis indicated that mRNA transcripts that annealed to a DARC probe under high stringency conditions were present in the spleen and other tissues of Duffy-negative individuals.21,22 As noted above, immunohistochemical staining revealed reactivity of anti-Fy6 with endothelial cells lining splenic sinusoids, bone marrow sinusoids, choroid plexus, and postcapillary venules in Duffy-negative as well as Duffy-positive individuals. Parallel biochemical studies demonstrated the presence of a high-affinity chemokine receptor (Kd ≈ 5 nM) with a binding specificity that included members of both C-X-C and C-C families, identical to that of DARC from the spleen of a Duffy individual.22 Nucleotide sequence analysis of the ORF from these patients failed to show evidence of a mutation in the coding sequences of the gene. As was observed by Chaudhuri et al, the genotype of the Duffy-negative African Americans tested was FY*B/FY*B.21 This was consistent with previous observations that Duffy-negative individuals of African descent produce anti-Fya antibodies, but not anti-Fyb.19 20
Based on the presence of an intact gene and a tissue-specific expression defect, it was postulated that the loss of expression in erythroid cells of Duffy-negative individuals could be due to a promoter/enhancer defect.20,108-112 Tournamille et al showed that Duffy-negative individuals have a point mutation in a consensus binding site for GATA-1, a transcription factor active in erythroid cells.110 This point mutation, located at −46, abolishes erythroid promoter activity in reporter gene assays.110 Further work is required to understand the molecular basis for maintenance of expression of DARC on endothelial cells and Purkinje cells, even in individuals who are Duffy negative.
The immunohistochemical and molecular genetic evidence suggest an important physiologic role for DARC. Although the selective pressure from malaria led to loss of DARC expression on RBCs (affording protection from P vivax malaria), nature's experiment selected a mechanism whereby expression of DARC was preserved on endothelial cells. This evolutionary outcome suggests that the function of DARC on endothelial cells is less dispensable than its function on RBCs.
Recently, the Duffy genotype of a rare Fy(a−b−) white individual was reported.113 This was the same individual (AZ) who produced anti-Fy3 described by Albrey et al.25 Analysis of the nucleotide sequence of amplification products from the DARC gene disclosed the presence of a 14-bp deletion that resulted in a frameshift mutation beginning at codon 98. AZ is apparently homozygous for this rare deletion. The mutation predicted the translation of a 118-amino acid residue polypeptide that would include the amino terminal extracellular domain, the first transmembrane spanning α-helix, and the first cytoplasmic loop. However, RBCs from this individual are devoid of serologically detectable Duffy antigen and presumably DARC is missing in other tissues as well. These studies were performed on blood cells that had been previously frozen, and neither current clinical information nor immunohistochemical information from nonerythroid tissue in this individual were obtainable. However, in the report by Albrey et al describing anti-Fy3 during AZ's third pregnancy, there is no mention of ill health. This suggests that a DARC-nullizygous phenotype may not be associated with adverse biologic consequences. Of course, biologic redundancy or compensatory mechanisms could obscure a defect in such individuals. It is hoped that a mouse nullizygous for the DARC homolog can be produced by targeted gene disruption to further explore the function of DARC. This was part of our rationale for cloning the murine DARC-homolog.
STRUCTURE-FUNCTION ANALYSIS OF DARC
Nucleotide sequence analysis of molecular clones of DARC genes from Fy(a+b−), Fy(a−b+), and Fy(a+b+) individuals in several laboratories showed that the Fya and Fyb alleles differ by a single base substitution in the second position of codon 44 (Fig 4) that encodes a glycine residue in Fya and an aspartic acid residue in Fyb.21,100 113-115 This polymorphism does not appear to have any biologic consequences.
Anti-Fy6 was shown to inhibit chemokine binding to DARC in a dose-dependent fashion whereas anti-Fy3 reagent did not alter binding at concentrations up to 100 μmol/L.116 At higher concentrations of anti-Fy3, some inhibition of chemokine binding to DARC was obtained.96,117 IL-8 did not inhibit the binding of anti-Fy6 to DARC on RBCs, indicating that the IL-8 binding site and the Fy6 epitope are distinct.118 The Fy6 and Fy3 epitopes were mapped using chimeric receptor proteins composed of complementary portions of DARC and IL-8RB, the chemoattractant receptor most closely related to DARC.116 Only chimeric receptors containing the amino terminal extracellular domain of DARC reacted with anti-Fy6 MoAbs, whether stably expressed in human cell lines or in insect cells using baculovirus. The Fy6 epitope was further localized by Wasniowska et al119 using a combinatorial peptide approach and site-directed mutagenesis. These investigators showed that two adjacent acidic amino acid residues, encoded by codons 25 (aspartic acid) and 26 (glutamic acid), are critical for anti-Fy6 binding, whereas the asparagine residues encoded by codons 18 and 29 are not involved (Fig 4). Furthermore, since mutation of these two residues eliminates two sites for the addition of oligosaccharide chains, it is likely that the Fy6 epitope is composed solely of peptide residues.
The epitope recognized by anti-Fy3 has also been mapped, although not to the same degree of molecular detail as Fy6.116 Immunofluorescent analysis of insect cells expressing chimeric receptors showed that the anti-Fy3 MoAb bound to constructs containing the three predicted extracellular loops of DARC, but not to constructs containing only the amino terminal extracellular domain.119 Anti-Fy3 did not bind to receptor chimeras that included the N-terminal extracellular domain and the first and second predicted extracellular loops. It was concluded that sequences necessary for the binding of anti-Fy3 are present in the third predicted extracellular loop of DARC (Fig 4).
Parallel studies localized the regions of the receptor involved in the binding of chemokines and the malarial Duffy binding ligands to this receptor.119 Analysis of a chimera expressing the N-terminal extracellular domain of DARC with the remainder of the heptahelical structure being that of IL-8RB (DARCe1/IL-RB) showed that it bound not only IL-8 and MGSA at high affinity, but also RANTES, a C-C chemokine that binds to DARC, but not IL-8RB. Moreover, the DARCe1/IL-8RB chimera bound the DARC-specific MGSA-E6A mutant with a Kd of 12 nM, mirroring the activity of intact DARC. Preliminary studies suggest that the 140-kD Duffy binding protein of P vivax also binds to the DARCe1/IL-8RB chimera (C. Chitnis, L.H. Miller, S. Peiper, personal communication, June 1995). Taken together, these findings highlight the significance of the amino terminal extracellular domain in the various biologic activities of DARC. Recent studies using DARC variants created by in situ mutagenesis indicate chemokine binding to DARC is dependent on the disulfide bond between a cysteine on the N-terminal extracellular domain and domain and a cysteine on the third extracellular loop.117
DARC IS EXPRESSED BY PURKINJE CELLS OF THE CEREBELLUM
Emerging evidence suggests that chemokines are involved in normal homeostatic processes in the central nervous system. Experiments using in situ hybridization localized rat mRNA that hybridizes to a human IL-8 probe in specific areas of the brain, including hippocampus and cerebellum.120 Human IL-8 has also been shown to enhance survival of rat hippocampal neurons in vitro.121 It is thought that IL-8 is synthesized and secreted by astrocytes.122 There are approximately 10-fold more astrocytes in the central nervous system than neurons and one of the functions of astrocytes is to “nurture” neurons by the secretion of neurotrophic factors. Until recently there were no reports on neurons expressing chemokine receptors.
Chaudhuri et al105 demonstrated the presence of mRNA in human brain that specifically hybridized with DARC cDNA. Interestingly, there were two species of mRNA observed: one of the predicted size, 1.2 kb, which was also detected in many other tissues tested; and one of 8.5 kb, which thus far has only been observed in brain. Although the nature of the 8.5-kb transcript has not yet been elucidated, the findings of DARC transcripts in the brain prompted our investigation of brain by immunohistochemistry.11,123 Anti-Fy6 was found to react specifically with Purkinje neurons of the cerebellum (Fig 6, see page 3083). Parallel experiments were done with an MoAb against IL-8 receptor B (IL-8RB) which reacted specifically with subsets of neurons in diverse regions of the brain and spinal cord.123 These included the hippocampus, dentate nucleus, pontine nuclei, locus coeruleus, and paraventricular nucleus in the brain and the anterior horn, interomediolateral cell column, and Clarke's column of the spinal cord. Like Purkinje neurons of the cerebellum that express DARC, the neurons in other areas of the brain and spinal cord that express the IL-8RB are projection neurons, bearing long axons that connect one neuronal region with another. Interneurons, which connect neurons within a region, did not react with either anti-Fy6 or the anti–IL-8RB antibody.
Further experiments were performed to confirm that anti-Fy6 reactivity with Purkinje cells was indeed caused by the presence of DARC. Membranes were isolated from cerebellar tissue at postmortem examination and these were analyzed for the presence of DARC by chemokine binding assays, chemokine cross-linking experiments, and immunblotting. Results obtained by each of these methods supported the immunohistochemical data and indicated that DARC is indeed expressed on Purkinje neurons of the cerebellum. What function DARC serves on Purkinje neurons and how this relates to its expression on RBCs and on endothelial cells remains to be determined.
THE RIDDLE OF DARC FUNCTION
Despite knowledge of structure-function relationships and tissue localization of DARC, the precise role of this receptor in normal and pathologic physiology remains uncertain. DARC does not appear to present chemokines in an active form to leukocyte receptors. Once bound to DARC expressed on the surface of erythrocytes, IL-8 did not induce effects in neutrophils associated with chemokine stimulation (K. Neote, S. Peiper, unpublished observations, September 1994).93 Human erythroleukemia (HEL) cells express DARC, but its function on these cells has not been elucidated.124 Although DARC on RBCs does not internalize ligand, there is evidence that DARC-transfectants can internalize ligands.22 It is possible that endothelial DARC internalizes ligands, thereby generating the chemotactic gradient essential for leukocyte attraction.
Based on what is known of other heptahelical membrane receptors, it seems likely that DARC transmits a signal across the membrane upon chemokine binding. However, DARC-dependent signal transduction has not yet been demonstrated. DARC does not appear to be coupled to a guanosine triphosphate binding protein (G-protein). It does not stimulate GTPase activity nor mediate calcium flux upon ligand binding; furthermore, DARC lacks a DRY motif in the second cytoplasmic loop that is characteristic of G-protein–coupled seven-membrane–spanning receptors.
To date, only a few other heptahelical receptors that lack DRY motifs have been cloned. They include the cAMP receptor in Dictostelium,bride of Sevenless (bos ) in drosophila, and the two seven-transmembrane–spanning proteins that harbor mutations in familial cases of Alzheimer's disease.125-128 The yeast homolog of the heptahelical protein mutated in Alzheimer's disease has been shown to be involved in signaling via the notch pathway.129 Thus, there is precedent for heptahelical receptors that signal through pathways independent of G-coupled proteins. It is possible that DARC interacts or “cross-talks” with other membrane components (which could differ depending on cell type) and thereby elicit tissue specific responses to ligand binding.
SUMMARY
The glycoprotein expressed on surface of erythrocytes initially known as the Duffy blood group antigen has been shown to be a receptor for the invasion of these cells by P vivax parasites. The parasite ligand that binds to the Duffy antigen has now been cloned and characterized. The region on the Duffy binding ligand of the parasite responsible for interaction with the Duffy antigen has also been identified. It is hoped that this molecule will useful as an immunogen to induce antibodies capable of blocking invasion of the parasite into the erythrocyte. Research on the Duffy binding ligand also provided a clue (a shared motif ) to the identification of the long-sought family of variant endothelial binding ligands that mediate attachment of P falciparum–infected erythrocytes to endothelial cells of postcapillary venules. If this attachment could be disrupted during the course of a malarial infection, it might lead to spleen-mediated parasite death. It might also lead to amelioration of cerebral malaria, a condition which in part is due to the blockade of cerebral venules by sequestered adherent parasites.
Recently, research on the Duffy antigen has taken on a new dimension. The Duffy antigen has been shown to be a multispecific heptahelical receptor for chemokines, expressed on RBCs, endothelial cells of postcapillary venules, and Purkinje cells of the cerebellum. The challenge now is to determine its function, both in immunobiology and neurobiology. Its capacity to bind chemoattractant cytokines and its expression on endothelial cells lining postcapillary venules are highly conserved across species, suggesting that this receptor subserves a critical function. This is supported by nature's experiment, the Duffy blood group negative phenotype, in which the genetic mechanism selected to remove expression on erythroid cells to protect against malarial infection, preserved expression on endothelial cells of postcapillary venules and splenic sinusoids. Although we have significant insight into structure-function relationships for the Duffy antigen/receptor for chemokines, its mechanism of signaling and its biologic function remain to be elucidated.
NOTE ADDED IN PROOF
Chaudhuri et al (Blood 89:701, 1997) recently described results of immunohistochemical staining with a polyclonal rabbit antibody (6615) that reacts with carbohydrate on the Duffy antigen. Reactivity was observed with endothelial cells of glomeruli, capillaries, vasa recta, and epithelial cells of collecting tubules in the kidney; reactivity was also observed with capillaries in the thyroid, and capillaries, large venules, and type I alveolar squamous cells of lung. Whether or not the carbohydrate epitope recognized by polyclonal rabbit antibody 6615 is also present on molecules other than the Duffy antigen remains to be determined.
ACKNOWLEDGMENT
We acknowledge the investigators in our laboratories including Zi-xuan Wang, Zhaohai Lu, and Haihong Guo. Collaborators at the University of Louisville include Alvin Martin and Victor Fingar and Stephen Slone. We also acknowledge Richard Horuk (Berlex, Richmond, CA), who has been a major collaborator, and Domanique Blanchard (Centre Regional de Transfusion Sanguine, Nantes, France) and Makoto Uchikawa (Japanese Red Cross Central Blood Center, Tokyo, Japan) for providing anti-Fy6 and anti-Fy3, respectively. Anti-Fy3 was obtained through Peter Byrne at the National Reference Laboratory (Rockville, MD). We also thank Beverly Kirkpatrick and Abby Carden for assistance in preparing the manuscript for this review.
Supported by a Merit Review Grant from the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, by the Agnes Brown Duggan Endowment for Oncologic Research, and the the Humana Endowment for Excellence.
Address reprint requests to Terence J. Hadley, MD, James Graham Brown Cancer Center, University of Louisville, 529 S Jackson St, Louisville, KY 40292.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal