Abstract
The human red cell Rh(D) antigen elicits the production of high-affinity IgG antibodies, which can prevent blood transfusion and cause hemolytic disease of the newborn. It has been known for 20 years that Rh(D) antibodies are among the most positively charged human serum IgGs. Analysis by IEF of 9 human anti-Rh(D) monoclonal antibodies showed that their isoelectric points (pI) (8.3 to 8.6) were also significantly higher than the average pI of serum IgGs (7.0 to 8.5). Sequencing of the anti-Rh(D) H and L chains cDNAs showed a preferential use of VH1 , VH3, JH6, and Vκ1 gene segments. The high pIs in IEF were correlated with a higher number of cationic amino acid residues in the H chain V regions without clustering in the complementary determining region. Computer analysis indicated that the germline VH used in anti-Rh(D) was selected among the most cationic segments available in the human VH repertoire or expressed in normal B cells. These results indicate that the selection of cationic VH segments may be an important early step in the formation of clinically relevant anti-Rh(D) and other red cell antibodies, possibly to facilitate epitope binding in the negatively charged red cell membrane environment.
AFTER THE ABO determinants, the Rh(D) antigen of the Rhesus blood groups is the most important human red cell antigen. The Rh(D) antigen is highly immunogenic in humans and elicits the production of high-affinity IgG antibodies. These antibodies can cause the destruction of Rh(D)+ red cells in transfused Rh(D)− individuals or the hemolytic disease of the Rh(D)+ fetus in Rh(D)− mother.1 It is known that the Rh antigens are carried on nonglycosylated integral membrane proteins, and several RH genes coding for membrane proteins of about 30 kD have been recently cloned.2 Although the fine structure of the Rh(D) epitopes is not yet defined, it is known that the Rh polypeptides are deeply buried in the red cell membrane with only short peptide loops, presumably carrying the Rh(D) epitopes, accessible on the red cell membrane.3
The Rh(D) antibodies have been extensively characterized serologically for 50 years.1,4 It was shown in 1969 that these antibodies were among the most cationic IgGs present in human serum.5 The exact role of this charge restriction has remained unclear, but it has been proposed1 that it could facilitate either their binding to putative acidic Rh(D) epitopes in agreement with the opposite charge hypothesis of specific antigen-antibody interactions,6 or rather their penetration in the layer of negative charges present on red cell membranes.7 Other cationic antibodies reacting with negatively charged DNA structures have been extensively studied in the last 10 years because of their implication in the development of nephritis associated with systemic lupus erythematosus (SLE).8 The cationic nature of DNA-specific antibodies could be related to the presence of positively charged amino acids in the antigen-binding complementary determining regions (CDRs) of the antibody V regions.9
The molecular basis of antibody structure and diversity is now known to be dependent on the recombination in pre-B lymphocytes, of various gene segments (VH , D, JH , VL , JL ) coding for the antigen-binding V regions of antibody H and L chains. In the last years, several human Rh(D) monoclonal antibodies (MoAbs) have been molecularly characterized. In an extensive study of 14 anti-Rh(D) MoAbs prepared from 7 different donors, Bye et al10 reported a restriction in the diversity of some of the gene segments used in anti-Rh(D) H chains. Indeed most H chains were coded by two rearranged VH segments of the VH3 and VH4 gene families and used JH6 segments. This study confirmed at the molecular level, the structural similarity among Rh(D) antibodies previously observed using VH gene family11 and idiotype-specific antisera.12 However, the restricted structural characteristics remained unclear. In this work, we have studied the relationship between Rh(D) antibody charge and V region primary structure. The cationic nature of Rh(D) IgG antibodies was confirmed using a library of 10 purified human Rh(D)-specific IgG MoAbs. Molecular characterization showed that the cationic nature of Rh(D) antibodies was correlated with the high pI of H chain V regions and we present evidence that the phenomenon is primarily due to the use in Rh(D) antibodies, of a limited set of VH gene segments selected among the most cationic VH gene segments available in the human VH germline repertoire and expressed in peripheral B cells.
MATERIALS AND METHODS
Cell lines.The 10 human-mouse heterohybridoma cell lines secreting human monoclonal anti-Rh(D) IgG were prepared by fusion of P3X63.Ag8.653 mouse myeloma cells with peripheral blood lymphocytes of two hyperimmunized Rh(D)− blood donors.13 The heterohybridoma cells were cloned by limiting dilution culture and expanded in Iscove's medium (Sigma Chemical Co, St Louis, MO) containing 10% fetal calf serum (Hyclone Laboratories Inc, Logan, UT), 50 U/mL penicillin (Sigma), and 50 μg/mL streptomycin (Sigma). Cells were cultured at 37°C in a humid incubator containing 10% CO2 . The anti-Rh(D)–containing culture supernatants were prepared by centrifugation of dense cultures, filtered on 0.45-μ membranes, supplemented with 0.01% NaN3 (Sigma), and stored at 4°C.
Isoelectric focusing.The IgG MoAbs were purified from 1 L of each culture supernatant by affinity chromatography on protein G-agarose (0.75-mL column) (GIBCO-BRL, Grand Island, NY). The IgGs were eluted with 0.1 mol/L glycine-HCL pH 2.6 and dialyzed against phosphate-buffered saline. The concentration of each MoAb solution was determined by human IgG-specific enzyme-linked immunosorbent assay as previously described.14
Isoelectric focusing (IEF) was performed in a thin-layer 5% polyacrylamide gel containing isoelectric point (pI) 3-10 ampholytes (Bio-Rad, Hercules, CA). Samples (3 to 4 μg of purified IgG MoAb or 20 μg of purified serum IgG) were focused in three steps: 15 minutes at 100 V, 15 minutes at 200 V, and 45 minutes at 400 V in a mini IEF cell (model 111, Bio-Rad). The gel was stained with Coomasie Brillant Blue (CBB) according to the manufacturer's instructions (Bio-Rad).
Amplification, cloning, and sequencing of anti-Rh(D) V genes.All primers used for cDNA synthesis and polymerase chain reaction (PCR) amplifications, except for the Universal Gamma Primer (UG), have been described previously.15 The 5′ primers contained an EcoRI restriction site to facilitate the cloning and are specific to a sequence in the leader peptide. The 3′ UG primer (5′AAGTGATCCTTGACCA-3′) is a universal primer capable of amplifying all four human gamma H chains. This primer is specific for a sequence located 30 base pair (bp) downstream from the JH segment in the first constant domain (CH1). Total RNA was prepared from 3 × 107 heterohybridoma cells by the guanidinium isothicyanate extraction procedure16 and purified by centrifugation through a CsCI cushion.17 First-strand cDNA synthesis was done at 42°C for 60 minutes in a 20-μL reaction volume containing: 1 × PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl pH 8.3, 1.5 mmol/L MgCl2 , and 0.01% gelatin) (Cetus-Perkin Elmer, Emeryville, CA), 200 μmol/L each deoxynucleotide triphosphate (dNTP), 5 pmol of the appropriate 3′ primers, 200 U of Moloney MuLV reverse transcriptase (GIBCO-BRL) and 100 ng of total RNA. For the PCR reaction, 80 μL of PCR mix containing 1 × PCR buffer, 50 pmol of the upstream and 45 pmol of the downstream primers and 2.5 U of Taq DNA Polymerase (Cetus-Perkin Elmer) were added to the 20-μL reverse transcriptase reaction. The reaction mix was overlaid with mineral oil and subjected to 30 rounds of amplification. The temperature used for the PCR were: denaturation at 94°C for 1 minute; annealing at 55°C for 2 minutes; extension at 72°C for 1 minute; with 1 minute ramp times. At the end of the 30 cycles, a 10 minute incubation at 72°C was done to complete elongation. An aliquot of the PCR products was analyzed by agarose gel electrophoresis in TBE buffer (90 mmol/L Tris-Borate, 2 mmol/L EDTA, pH 8.0) and stained with ethidium bromide. The amplified VH and VL cDNAs were recovered from the gel and purified with the GeneClean II kit (BIO 101, Inc, Vista, CA). The cDNAs were digested with EcoRI, phosphorylated and ligated in pBluescript SK + vector (Stratagene, La Jolla, CA) and used to transform JM109 competent Escherichia coli cells. Recombinant plasmid DNA was alkali-denatured and sequenced with the T7 sequencing kit (Pharmacia, Uppsala, Sweden). At least two clones from separate PCR amplifications were sequenced for each VH and VL .
Computer analysis of DNA sequences.The closest rearranged V genes were determined by comparing H and L chain variable segments to the ones present in GenBank (release 89.0) and EMBL data bases (release 42.0) using the Blast algorithm at the National Center for Biology Information (NCBI, Bethesda, MD) server.18 Germline VH segment identity was established by comparing VH and VL segments to the compiled germline data base (V base) of Cook and Tomlinson.19 All other computer analyses of DNA sequences were performed using the Genetic Computer Group (GCG, University of Wisconsin, Madison) sequence analysis software (version 8.0.1).20 The isoelectric points of the H and L chain V regions were calculated with the Isoelectric program without including the leader peptide.
RESULTS
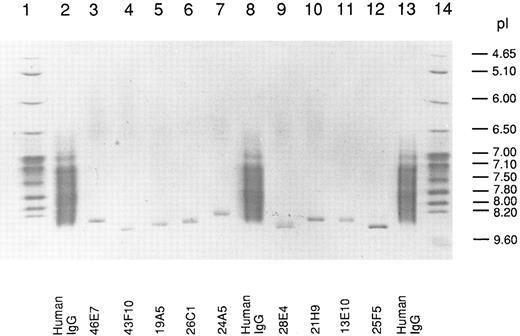
Isoelectric point of human anti-Rh(D) IgG MoAbs.We used in this work a library of 10 human-mouse anti-Rh(D)–secreting heterohybridoma cell lines prepared in two fusion experiments done with the PBLs of two Rh(D)-immunized donors (8 and 2 cell lines, respectively) (see Table 1). The Rh(D) specificity of the 10 IgG MoAbs was confirmed by standard red cell serology using panels of phenotyped red cells from our blood bank. The MoAbs were purified from culture supernatants by protein G affinity chromatography and their pI was determined by IEF in parallel with normal human serum polyclonal IgG. A photograph of the stained gel is shown in Fig 1. All the MoAbs (except 13D2, which could not be purified) yielded unique IEF bands with pI between 8.3 and 8.6. As expected, polyclonal serum IgG produced a smear in the 7.0 to 8.5 pI range. Among the six IgG1 (κ) present in the anti-Rh(D) MoAbs library, the decreasing order of relative pI observed by IEF was: 43F10/25F5-28E4-19A5-21H9-46E7. This result showed that the cationic nature of human Rh(D) antibodies first described in polyclonal anti-Rh(D) sera5 was also observed with all tested anti-Rh(D) IgG MoAbs prepared in vitro. Antibody isotyping showed that the anti-Rh(D) MoAbs were of the common IgG1 (6 MoAbs) and IgG3 (3 MoAbs) subclasses and used both κ (8 MoAbs) and λ (2 MoAbs) light chains (Table 1) ruling out the possibility that the relative cationic charge might be due to a restriction in C region diversity. Also deglycosylation of some anti-Rh(D) MoAbs by treatment with N-glycosidase F did not significantly reduce the pIs observed in IEF indicating that glycosylation was not responsible for the overall cationic charge (data not shown).
Germline and Rearranged VH and VL Gene Segments Used in Anti-Rh(D) H and L Chains
| Family . | Antibody* . | Isotype . | Probable† Germline V Gene . | Nucleotide Differences . | Next closest† Germline V Gene . | Nucleotide Differences . | Closest† Rearranged V Gene . | Nucleotide Differences . |
|---|---|---|---|---|---|---|---|---|
| VH1 | 25F5 | IgG1/k | V1-2 | 8 | DP-8 | 9 | 20P3 fetal | 7 |
| VH1 | 13E10 | IgG1/λ | V1-2 | 12 | DP-8 | 13 | 20P3 fetal | 13 |
| VH1 | 21H9 | IgG1/k | DP-25 | 11 | VI-3 | 16 | LE 1-42 fetal | 11 |
| VH1 | 28E4 | IgG1/k | DP-7 | 14 | HG3 | 15 | H20C3H fetal | 16 |
| VH3 | 24A5 | IgG3/λ | hv3019b9 | 5 | P6 | 7 | FL13-48 fetal | 5 |
| VH3 | 26C1 | IgG3/k | hv3019b9 | 5 | P6 | 7 | FL13-48 fetal | 5 |
| VH3 | 19A5 | IgG1/k | hv3019b9 | 17 | P6 | 19 | FL13-48 fetal | 17 |
| VH3 | 43F10 | IgG1/k | hv3019b9 | 16 | P6 | 18 | FL13-48 fetal | 16 |
| VH3 | 13D2 | IgG3/k | hv3019b13 | 9 | DP-46 | 10 | M74 fetal | 9 |
| VH3 | 46E7 | IgG1/k | 3-21 | 16 | 3-48 | 20 | E54 fetal | 13 |
| VkI | 26C1 | IgG3/k | L11 | 10 | A30 | 21 | 2E7 anti-platelet | 18 |
| VkI | 13D2 | IgG3/k | 02 | 8 | L11 | 23 | 15a anti-i | 5 |
| VkI | 21H9 | IgG1/k | 02 | 12 | L11 | 27 | 15a anti-i | 12 |
| VkI | 43F10 | IgG1/k | 02 | 13 | L11 | 29 | 15a anti-i | 13 |
| VkI | 46E7 | IgG1/k | L12a | 6 | HK102 | 10 | I.26 | 6 |
| VkI | 25F5 | IgG1/k | L12a | 15 | HK102 | 19 | I.26 | 10 |
| VkIII | 28E4 | IgG1/k | DPK21 | 2 | Vg | 19 | Z18330‡ | 2 |
| VkIII | 19A5 | IgG1/k | Humkv325 | 7 | Humvk305 | 12 | T15-42 | 6 |
| VλII | 13E10 | IgG1/λ | DPL-11 | 2 | DP-L10 | 16 | L25295‡ | 2 |
| VλIV | 24A5 | IgG3/λ | IGLV3S1 | 12 | Vλ III.1 | 73 | E51 | 10 |
| Family . | Antibody* . | Isotype . | Probable† Germline V Gene . | Nucleotide Differences . | Next closest† Germline V Gene . | Nucleotide Differences . | Closest† Rearranged V Gene . | Nucleotide Differences . |
|---|---|---|---|---|---|---|---|---|
| VH1 | 25F5 | IgG1/k | V1-2 | 8 | DP-8 | 9 | 20P3 fetal | 7 |
| VH1 | 13E10 | IgG1/λ | V1-2 | 12 | DP-8 | 13 | 20P3 fetal | 13 |
| VH1 | 21H9 | IgG1/k | DP-25 | 11 | VI-3 | 16 | LE 1-42 fetal | 11 |
| VH1 | 28E4 | IgG1/k | DP-7 | 14 | HG3 | 15 | H20C3H fetal | 16 |
| VH3 | 24A5 | IgG3/λ | hv3019b9 | 5 | P6 | 7 | FL13-48 fetal | 5 |
| VH3 | 26C1 | IgG3/k | hv3019b9 | 5 | P6 | 7 | FL13-48 fetal | 5 |
| VH3 | 19A5 | IgG1/k | hv3019b9 | 17 | P6 | 19 | FL13-48 fetal | 17 |
| VH3 | 43F10 | IgG1/k | hv3019b9 | 16 | P6 | 18 | FL13-48 fetal | 16 |
| VH3 | 13D2 | IgG3/k | hv3019b13 | 9 | DP-46 | 10 | M74 fetal | 9 |
| VH3 | 46E7 | IgG1/k | 3-21 | 16 | 3-48 | 20 | E54 fetal | 13 |
| VkI | 26C1 | IgG3/k | L11 | 10 | A30 | 21 | 2E7 anti-platelet | 18 |
| VkI | 13D2 | IgG3/k | 02 | 8 | L11 | 23 | 15a anti-i | 5 |
| VkI | 21H9 | IgG1/k | 02 | 12 | L11 | 27 | 15a anti-i | 12 |
| VkI | 43F10 | IgG1/k | 02 | 13 | L11 | 29 | 15a anti-i | 13 |
| VkI | 46E7 | IgG1/k | L12a | 6 | HK102 | 10 | I.26 | 6 |
| VkI | 25F5 | IgG1/k | L12a | 15 | HK102 | 19 | I.26 | 10 |
| VkIII | 28E4 | IgG1/k | DPK21 | 2 | Vg | 19 | Z18330‡ | 2 |
| VkIII | 19A5 | IgG1/k | Humkv325 | 7 | Humvk305 | 12 | T15-42 | 6 |
| VλII | 13E10 | IgG1/λ | DPL-11 | 2 | DP-L10 | 16 | L25295‡ | 2 |
| VλIV | 24A5 | IgG3/λ | IGLV3S1 | 12 | Vλ III.1 | 73 | E51 | 10 |
21H9 and 19A5 MoAbs were derived of the peripheral blood lymphocytes (PBLs) of donor LA and the other MoAbs of the PBLs of donor AR.
References for germline and rearranged genes are as follows: Germline VH: VI-2 and VI-321; DP-25, DP7, DP-8 and DP-4622; hv3019b9 and hv3019b1323; 3-21 and 3-4824; HG325. Rearranged VH 20P326; LE 1-4227; H20C3H28; FL13-4829; M74,30 and E5431. Germline VL: L1132; 0233; L12a and A3034; DPK2135; Humkv32536; HK10237; Humvk30538; Vg39; DPL-11 and DPL-1040; IGLV3S141 and VλIII.1.42 Rearranged VL: 2E743; 15a44; I.2645; T15-4246; L25295,47 and E51.48
Genbank accession number.
Isoelectric focusing of anti-Rh(D) IgGs. After electrophoresis, the gel was stained with CBB and photographed. The migration of pI standard proteins is shown in lanes 1 and 14 and of polyclonal human IgG in lanes 2, 8, and 13. The position of the 9 anti-Rh(D) MoAbs is shown at the bottom of the gel.
Isoelectric focusing of anti-Rh(D) IgGs. After electrophoresis, the gel was stained with CBB and photographed. The migration of pI standard proteins is shown in lanes 1 and 14 and of polyclonal human IgG in lanes 2, 8, and 13. The position of the 9 anti-Rh(D) MoAbs is shown at the bottom of the gel.
These results suggested that the high pI of anti-Rh(D) IgGs was dependent of the primary amino acid (a.a.) sequence of V regions.
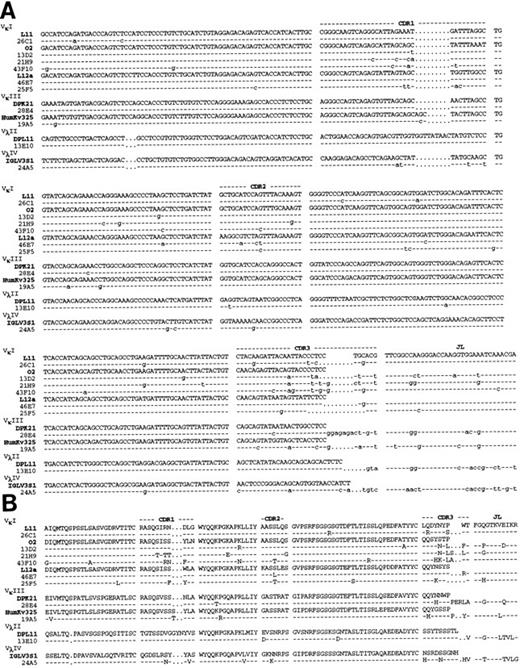
Molecular sequencing of anti Rh(D) V regions.H and L chain V region cDNAs were amplified by PCR, cloned and sequenced. The confirmed nucleotide and deduced a.a. sequences of the 10 anti-Rh(D) MoAbs are shown in Fig 2 (H chains) and Fig 3 (L chains). Germline VH , Vκ , and Vλ gene segment identification was done by comparison with the database compiled by Cook and Tomlinson.19 Results are listed in Table 1. Definitive identification of the germline VH segment was not possible because 8 MoAbs used VH segments highly homologous to two germline VH segments (difference of 1 or 2 bases). The germline D, JH , and JL segments used in the 10 MoAbs are listed in Table 2.
Nucleotide (A) and deduced amino acid (B) sequences of anti-Rh(D) H chain V regions. The H chains are classified according to VH families and germline VH used (V1-2, DP-25, DP7, hv3019b9, hv3019b13, 3-21). Dashes indicate identity. The JH segment is a consensus sequence. References for germline genes are in Table 1. Genbank accession numbers for anti-Rh(D) H chains are as follows: 25F5:U43758, 13E10:U43762, 21H9: U43760, 28E4:U43756, 24A5:U43759, 26C1:U43757, 19A5:U43761, 43F10:U43755, 13D2:U43763, and 46E7:U43754.
Nucleotide (A) and deduced amino acid (B) sequences of anti-Rh(D) H chain V regions. The H chains are classified according to VH families and germline VH used (V1-2, DP-25, DP7, hv3019b9, hv3019b13, 3-21). Dashes indicate identity. The JH segment is a consensus sequence. References for germline genes are in Table 1. Genbank accession numbers for anti-Rh(D) H chains are as follows: 25F5:U43758, 13E10:U43762, 21H9: U43760, 28E4:U43756, 24A5:U43759, 26C1:U43757, 19A5:U43761, 43F10:U43755, 13D2:U43763, and 46E7:U43754.
Nucleotide (A) and deduced amino acid (B) sequences of anti-Rh(D) L chain V regions. The L chains are classified according to isotype and to VL families and germline segments used (L11, 02, L12a, DPK21, HumKv325, DPL11, IGLV3S1). Dashes indicate identity. The JL segment is a consensus sequence. References for germline genes are in Table 1. Genbank accession numbers for anti-Rh(D) L chains are as follows: 26C1:U43767, 13D2:U43773, 21H9:U43770, 43F10:U43765, 46E7:U43764, 25F5:U43768, 28E4:U43766, 19A5:U43771, 13E10:U43772, and 24A5:U43769.
Nucleotide (A) and deduced amino acid (B) sequences of anti-Rh(D) L chain V regions. The L chains are classified according to isotype and to VL families and germline segments used (L11, 02, L12a, DPK21, HumKv325, DPL11, IGLV3S1). Dashes indicate identity. The JL segment is a consensus sequence. References for germline genes are in Table 1. Genbank accession numbers for anti-Rh(D) L chains are as follows: 26C1:U43767, 13D2:U43773, 21H9:U43770, 43F10:U43765, 46E7:U43764, 25F5:U43768, 28E4:U43766, 19A5:U43771, 13E10:U43772, and 24A5:U43769.
Identification of D and J Gene Segments Used in the H and L Chains of Anti-Rh(D) MoAbs
| Antibody . | D Segment* . | JH Segment* . | JL Segment* . |
|---|---|---|---|
| 25F5 | D21/9 | JH3a | Jκ1 |
| 13E10 | DN4 | JH6b | Jλ2/Jλ3† |
| 21H9 | DLR4 | JH6c | Jκ4 |
| 28E4 | D21/9 | JH5b | Jκ4 |
| 24A5 | DLR3 | JH6c | JλL |
| 26C1 | DA4 | JH6c | Jκ1 |
| 19A5 | DK4 | JH6b | Jκ2 |
| 43F10 | XP′1 | JH6a/b‡ | Jκ1 |
| 13D2 | DM5 | JH6c | Jκ3 |
| 46E7 | DXP1, DHQ52 | JH6c | Jκ1 |
| Antibody . | D Segment* . | JH Segment* . | JL Segment* . |
|---|---|---|---|
| 25F5 | D21/9 | JH3a | Jκ1 |
| 13E10 | DN4 | JH6b | Jλ2/Jλ3† |
| 21H9 | DLR4 | JH6c | Jκ4 |
| 28E4 | D21/9 | JH5b | Jκ4 |
| 24A5 | DLR3 | JH6c | JλL |
| 26C1 | DA4 | JH6c | Jκ1 |
| 19A5 | DK4 | JH6b | Jκ2 |
| 43F10 | XP′1 | JH6a/b‡ | Jκ1 |
| 13D2 | DM5 | JH6c | Jκ3 |
| 46E7 | DXP1, DHQ52 | JH6c | Jκ1 |
References for the germline genes are as follows: D segments: DN4, DLR4, DLR3, DA4, DK4, XP′1, and DXP149; D21/950; DM551; DHQ5252. JH segments: JH3a and JH6a52; JH6b, JH6c and JH5b are consensus sequences derived from rearranged genes.53 JL segments: Jκ54; Jλ.55
Same gene.
The 43F10 JH segment is highly homologous to both JH6a and JH6b.
For the 10 H chains, a total of 6 different VH gene segments were used. Two of these VH segments were used in four (hV3019b9) and two (VI-2) H chains. The germline VH were frequently mutated in anti-Rh(D) (average of 7.7 replacement mutations) particularly in the CDRs (CDR/framework [FR] ratio of 1.33). D gene segment usage was highly diversified with only one segment (D21/9) used in two MoAbs. One MoAb (46E7) used a tandem of 2-D segments (DPX1 and DHQ52) in the same orientation and consequently exhibited a CDR3 region of unusual length (26 amino acids). Eight of the 10 H chains used JH6b and JH6c polymorphic forms of the JH6 gene family whereas the two others used JH3a and JH5b gene segments.
There was more diversity in the antibody L chains due to the fact that both κ (8 MoAbs) and λ (2 MoAbs) chains were used. Three VκI gene segments were used in 6 MoAbs with the other four using different Vκ and Vλ gene segments. There was no evidence of preferential use of unique pairs of VH and VL gene segments in several antibodies. Four antibodies used Jκ1 gene segments but a total of 6 different JL segments were used in the 10 light chains. The germline VL gene segments used were less frequently mutated than the VH gene segments (average of 3.2 replacement mutations). Three VL segments showed only one replacement mutation with two others with only two. Also there was no clustering of these mutations in the CDRs (CDR/FR ratio of 1.1).
Overall, this analysis showed a preferential use of some V gene segments in anti-Rh(D). This was most clearly seen in H chains, which frequently used the hv3019b9 VH (4/10) and JH6 (8/10) gene segments. This result confirmed the ones of Bye et al10 who reported the preferential use in anti-Rh(D) of some VH and JH6 gene segments.
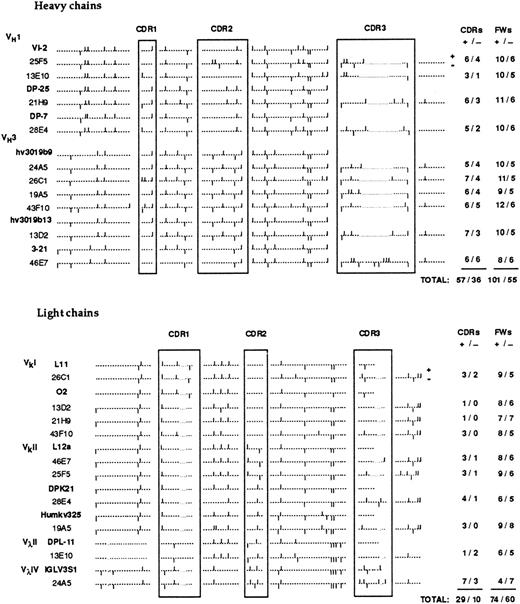
Molecular basis for the high pI of anti-Rh(D).To correlate antibody cationic charge and primary V region structure, we first analyzed the distribution of cationic and anionic amino acids in the H and L chain V regions. The result is illustrated in Fig 4. Overall, the antibody V regions contained far more cationic (261) than anionic (161) a.a. residues (+/− ratio of 1.62). This ratio is significantly higher than the one (1.22) calculated from the average content in charged a.a. of more than 1,000 antibody V regions (from summary tables in ref. 56) indicating that the V region primary sequence contributes significantly to the cationic charge of anti-Rh(D) compared to average serum IgGs. Of the 100 additional cationic residues, 67 were located in the H chains and these residues were not localized predominantly in the CDRs (+/− ratio of 1.58 in the CDRs and 1.83 in FRs). However, a significant enrichment of cationic residues was observed in the CDRs of L chains (+/− ratio of 2.90 in CDRs and 1.23 in FRs). Thus the high pI of these human anti-Rh(D) (Fig 1) could be related to the presence of more positively charged amino acids in the whole H chains V regions and to a lesser extent in the L chain CDRs.
Distribution of cationic and anionic amino acids in the V regions of anti-Rh(D) H and L chains. The position of charged residues is shown by bars above (+) and below (−) the sequence line. The numbers of cationic and anionic amino acids in the CDRs and FRs are indicated at the right.
Distribution of cationic and anionic amino acids in the V regions of anti-Rh(D) H and L chains. The position of charged residues is shown by bars above (+) and below (−) the sequence line. The numbers of cationic and anionic amino acids in the CDRs and FRs are indicated at the right.
Since the contribution of the charged amino acid residues to protein pI varies according to their pKa, the pI of the different V regions and isolated VH and VL fragments was determined by computer analysis. The results are listed in Table 3. The analysis first confirmed that the average pI of anti-Rh(D) H chain V regions (10.05 ± 0.31) is much higher than the one of L chain V regions (8.84 ± 1.64). For six of the 10 H chains, the calculated pI of the VH segment alone was higher than the one of the whole V region indicating that the selected D and JH gene segments coding for CDR3 and FR4 H chain regions did not always contribute to the high pI of anti-Rh(D). The germline VH segments used in anti-Rh(D) H chains showed evidence of somatic mutations (see Table 1) suggesting that the basic a.a. residues could be derived from the somatic mutation process. However, the average pI of the anti-Rh(D) VH segments (10.05) was only slightly higher than the one of the germline VH segments (9.96) indicating that the somatic mutation process was not a predominant factor in the formation of cationic H chains. Furthermore, the pIs of 4 anti-Rh(D) VH segments were equal or lower than the ones of the corresponding germline VH segments. The situation was clearly different for L chains. The calculated pI of the VL fragments was lower than the one of the whole VL regions in all 10 MoAbs indicating that CDR3 and FR4 residues coded by JL gene segments contribute significantly to L chain cationic charge. Mutations in the VL gene segments during immune response did not appear to influence much VL charge since in seven out of 10 cases, the pI of the germline VL segment used was unchanged or lower than the one of the corresponding anti-Rh(D) VL fragment. Overall, this analysis showed the predominant role of germline VH gene segment selection for H chains and to a lesser extent of JL gene segment selection for L chains in determining the overall cationic change of human Rh(D) antibodies.
Calculated Isoelectric Points of Anti-Rh (D) H and L Chains
| Antibody . | Calculated Isoelectric Points3-150 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | Heavy Chain . | Light Chain . | . | . | . | . | ||||
| . | VDJ . | VH . | Germline VH . | VJ . | VL . | Germline VL . | . | . | . | . |
| 25F5 | 10.04 | 10.35 | 10.18 | 9.80 | 8.07 | 8.07 | ||||
| 13E10 | 10.24 | 10.05 | 10.18 | 5.72 | 4.82 | 5.72 | ||||
| 21H9 | 10.02 | 10.17 | 10.39 | 8.07 | 4.77 | 8.07 | ||||
| 28E4 | 10.28 | 10.31 | 9.88 | 9.45 | 6.41 | 8.07 | ||||
| 24A5 | 10.02 | 9.96 | 9.92 | 6.23 | 4.36 | 4.96 | ||||
| 26C1 | 10.11 | 10.33 | 9.92 | 10.22 | 9.44 | 8.07 | ||||
| 19A5 | 10.13 | 9.92 | 9.92 | 9.37 | 8.41 | 6.45 | ||||
| 43F10 | 9.90 | 10.09 | 9.92 | 10.38 | 9.48 | 8.07 | ||||
| 13D2 | 10.48 | 9.92 | 9.92 | 9.36 | 8.07 | 8.07 | ||||
| 46E7 | 9.28 | 9.41 | 9.34 | 9.86 | 8.07 | 8.07 | ||||
| Mean ± SD | 10.05 ± 0.31 | 10.05 ± 0.28 | 9.96 ± 0.27 | 8.84 ± 1.64 | 7.19 ± 1.94 | 7.36 ± 1.19 | ||||
| Antibody . | Calculated Isoelectric Points3-150 . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| . | Heavy Chain . | Light Chain . | . | . | . | . | ||||
| . | VDJ . | VH . | Germline VH . | VJ . | VL . | Germline VL . | . | . | . | . |
| 25F5 | 10.04 | 10.35 | 10.18 | 9.80 | 8.07 | 8.07 | ||||
| 13E10 | 10.24 | 10.05 | 10.18 | 5.72 | 4.82 | 5.72 | ||||
| 21H9 | 10.02 | 10.17 | 10.39 | 8.07 | 4.77 | 8.07 | ||||
| 28E4 | 10.28 | 10.31 | 9.88 | 9.45 | 6.41 | 8.07 | ||||
| 24A5 | 10.02 | 9.96 | 9.92 | 6.23 | 4.36 | 4.96 | ||||
| 26C1 | 10.11 | 10.33 | 9.92 | 10.22 | 9.44 | 8.07 | ||||
| 19A5 | 10.13 | 9.92 | 9.92 | 9.37 | 8.41 | 6.45 | ||||
| 43F10 | 9.90 | 10.09 | 9.92 | 10.38 | 9.48 | 8.07 | ||||
| 13D2 | 10.48 | 9.92 | 9.92 | 9.36 | 8.07 | 8.07 | ||||
| 46E7 | 9.28 | 9.41 | 9.34 | 9.86 | 8.07 | 8.07 | ||||
| Mean ± SD | 10.05 ± 0.31 | 10.05 ± 0.28 | 9.96 ± 0.27 | 8.84 ± 1.64 | 7.19 ± 1.94 | 7.36 ± 1.19 | ||||
For the six IgG1 (κ) anti-Rh(D) included in our study, we calculated the decreasing order of pI for V regions (average of H and L chains pIs) (Table 3). The order obtained, 43F10-25F5-28E4-19A5- 46E7-21H9, is identical with the one observed during the IEF analysis of the antibody molecules (Fig 1) except for the two last positions that are inversed. Such a good correlation supports the conclusion that the high pI of the V region primary a.a. sequence is responsible for the restricted cationic charge of anti-Rh(D) IgGs.
Since germline VH but not VL gene segments appear to be of primary importance for the positive charge restriction in Rh(D) antibodies, we next compared the pI of the germline VH and VL segments used in these antibodies to the ones of all the segments reported in the human repertoire.19 After computer analysis, the germline segments were classified according to pI increments. The result (Table 4) showed that the germline VH and VL segments could be distributed in 8 groups over a wide range of pI from below 8.00 to more than 10.00 for VH and from below 4.20 to more than 9.00 for VL . In the case of VH segments, it is known that some segments are more frequently used in human antibodies and also included in Table 4 is the pI distribution of the 28 germline VH segments most frequently expressed in normal B cells along with their usage frequency.57 This distribution is not significantly different from the one of the germline repertoire and the usage frequency indicated the use by normal B cells, of VH segments with a wide range of pI. The VH segments in some of the pI intervals were used more frequently because of the presence in these groups of the most frequently used individual VH segments such as DP47 (16.9%, pI = 8.65) and DP49 (8.5%, pI = 9.87).60 The distribution obtained for the six VH segments used in anti-Rh(D) was biased because the 6 VH segments were present among the three more basic VH groups. Thus the 6 anti-Rh(D)VH were selected among the 25 most cationic VH segments out of 51 present in the germline repertoire, and among the 15 most cationic segments out of 28 expressed in peripheral B cells. It should be pointed out that this group of 15 most cationic VH segments represented a total usage frequency of less than 50% in normal B cells. The probability that such a biased pI distribution of anti-Rh(D) VH could be obtained by random selection in the germline repertoire is low (P = .02). Five out of six (except DP-25) anti-Rh(D) VH were among the 28 VH segments expressed in normal B cells but these VH segments were not among the most frequently used VH segments since their total usage frequency was only 11.2%. In the case of the L chains and in agreement with the wider ditribution of the pIs of anti-Rh(D) VL segments (see Table 3), no biased use of cationic VL segments was observed in anti-Rh (D) L chains.
Comparative Distribution of pI of VH and VL Segments Available in the Germline Repertoire and Used in Peripheral B Cells and Anti-Rh(D)
| pI Interval . | No. of VH Segments in pI Interval . | ||
|---|---|---|---|
| . | Germline Repertoire . | Peripheral B Cells Usage (% of total)4-150 . | Anti-Rh(D)4-151 . |
| VH | |||
| >10.00 | 5 | 1 (2.8%) | 2 (DP-25, VI-2) |
| 9.66-9.99 | 15 | 10 (36.5%) | 3 (hv3019b9, DP-7, and hv3019b13) |
| 9.33-9.65 | 5 | 4 (8.4%) | 1 (3-21) |
| 9.00-9.32 | 10 | 5 (12.6%) | 0 |
| 8.66-8.99 | 4 | 1 (7.0%) | 0 |
| 8.33-8.65 | 5 | 4 (28.1%) | 0 |
| 8.00-8.32 | 3 | 2 (2.8%) | 0 |
| <8.00 | 4 | 1 (1.4%) | 0 |
| VL | |||
| >9.00 | 2 | — | 0 |
| 8.40-8.99 | 3 | — | 0 |
| 7.80-8.39 | 10 | — | 4(L11, L12a, 02, and DPK21) |
| 7.20-7.79 | 4 | — | 0 |
| 6.60-7.19 | 5 | — | 0 |
| 6.00-6.59 | 8 | — | 1 (Humkv325) |
| 5.40-5.99 | 5 | — | 1 (DPL11) |
| 4.80-5.39 | 8 | — | 1 (IGLV3S1) |
| 4.20-4.79 | 10 | — | 0 |
| <4.20 | 2 | — | 0 |
| pI Interval . | No. of VH Segments in pI Interval . | ||
|---|---|---|---|
| . | Germline Repertoire . | Peripheral B Cells Usage (% of total)4-150 . | Anti-Rh(D)4-151 . |
| VH | |||
| >10.00 | 5 | 1 (2.8%) | 2 (DP-25, VI-2) |
| 9.66-9.99 | 15 | 10 (36.5%) | 3 (hv3019b9, DP-7, and hv3019b13) |
| 9.33-9.65 | 5 | 4 (8.4%) | 1 (3-21) |
| 9.00-9.32 | 10 | 5 (12.6%) | 0 |
| 8.66-8.99 | 4 | 1 (7.0%) | 0 |
| 8.33-8.65 | 5 | 4 (28.1%) | 0 |
| 8.00-8.32 | 3 | 2 (2.8%) | 0 |
| <8.00 | 4 | 1 (1.4%) | 0 |
| VL | |||
| >9.00 | 2 | — | 0 |
| 8.40-8.99 | 3 | — | 0 |
| 7.80-8.39 | 10 | — | 4(L11, L12a, 02, and DPK21) |
| 7.20-7.79 | 4 | — | 0 |
| 6.60-7.19 | 5 | — | 0 |
| 6.00-6.59 | 8 | — | 1 (Humkv325) |
| 5.40-5.99 | 5 | — | 1 (DPL11) |
| 4.80-5.39 | 8 | — | 1 (IGLV3S1) |
| 4.20-4.79 | 10 | — | 0 |
| <4.20 | 2 | — | 0 |
Data from Brezinschek et al.57
VH name in parentheses.
DISCUSSION
In a previous study, Bye et al10 reported the preferential use of some VH3 and VH4 and of JH6 gene segments in human Rh(D) IgG antibodies. They also showed evidence of somatic mutation and repertoire shift during the maturation of Rh(D) immune response. The analysis of our 10 Rh(D) IgG antibodies confirms and extends these results. The frequent use of the hv3019B9 VH3 gene segment in anti-Rh(D)10 was also observed in 4 of our 10 MoAbs as well as the preferential use of JH6 (8/10) gene segments. However, we did not find VH4 gene segments in our MoAbs. Rather four MoAbs used 3 segments (V1-2, DP25, DP7) of the VH1 gene family. Use of the DP25 segment has already been reported in another anti-Rh(D).58 Furthermore a VH5-encoded anti-Rh(D) has recently been reported.59 Together these results indicate that the limited set of VH gene segments used in IgG anti-Rh(D) can at least be derived from four VH gene families. The functional basis for the VH restriction has remained unclear. However, the high diversity of anti-Rh(D) CDRs suggested that the VH restriction could be important for some structural characteristics of the anti-Rh(D) other than Rh(D) epitope binding per se.
The cationic charge of human anti-Rh(D) IgGs has been known for 20 years (5) and is routinely used to partially purify the prophylaxis Rh(D) IgG preparations from human plasma by anion exchange chromatography. In this work, we first showed that this characteristic was also observed in all tested human anti-Rh(D) IgG MoAbs confirming the extent of this charge restriction. Analysis of V region sequences showed that the high pI of all anti-Rh(D) was correlated with the presence of cationic H but not necessarily L chain V regions. The high pI of H chains was not primarily derived from somatic mutations or the formation of cationic CDR3 regions since there was only a slight difference (0.09) between the average pIs of the used germline VH segments and of the whole H chain V regions. A significant increase (1.48) was, however, noted in L chains by the recombination of VL segments with cationic JL segments. The finding that 6 out of 6 germline VH segments used in our anti-Rh(D) were present in the most cationic half of VH segments present in the germline repertoire or expressed in normal B cells indicates that the selection was not random. Inclusion in this analysis of 6 other germline VH segments (DP49, H11, DP-53, VH4-21, V71-4, V2-1) previously observed in other anti-Rh(D) IgGs10,58-60 supports the finding by showing that 10 out of 12 anti-Rh(D) VH are in the most cationic half of germline VH segments. Taking into account that some of these segments have been used in several anti-Rh(D), 24 out of 26 previously sequenced anti-Rh(D) IgGs are coded by VH segments selected from the most cationic half. No such biased use of cationic VL segments was noted in our MoAbs and in the previously reported anti-Rh(D) L chains,10 which are frequently coded by the rather anionic IGLV3S1 and Humkv325 VL segments used in our 24A5 and 19A5 MoAbs. Although the more cationic VH segments were not more frequently expressed in normal B cells, it could be argued that they are also more frequently used in other specific human immune responses. Additional analysis of H chains of many human MoAbs reacting with structures unrelated to blood group antigens61-63 showed that most of them (49 out of 64 H chains) were actually coded by VH segments selected among the less cationic half of germline VH segments (pI < 9.32). This analysis confirms a recently published compilation study,64 which showed that human antibodies produced during active immune responses were frequently (51%) coded by a limited set of six germline VH segments. These segments are among those frequently used in peripheral B cells57 and have an average pI of only 9.10. These results are in agreement with the average pI of serum IgG and indicates that the selection of cationic germline VH segments is probably an important mechanism in the generation of high-affinity anti-Rh(D) V regions.
The finding that the anti-Rh(D) basic amino acid residues are not clustered in CDRs strongly suggests that the functional role of cationic restriction is not related to the binding to Rh(D) epitopes. That interacting surfaces of antibodies and antigens may have opposite charges is well established6 and supported recently by the sequence analysis of several cationic anti-DNA MoAbs which showed the presence in CDRs of several arginine residues for ionic bonding to anionic DNA.9 On the other hand, red blood cells are known to be surrounded by a cloud of negative charges composed of sialic acid to prevent spontaneous cell-cell adhesion in circulation.7 The cationic charge of human anti-Rh(D) may thus be required for penetration of the antibody in this environment to allow the binding to the Rh(D) antigen, which is known to be located close to membrane lipids. Genetic engineering of anti-Rh(D) MoAbs to reduce or increase overall antibody charge without affecting CDRs structure would permit to test this hypothesis. Such a mechanism would also indicate that the high affinity of human immune anti-Rh(D) IgGs, which is always measured with intact red cells could result from the synergizing effects of Rh(D)-specific paratope binding and of general anti-Rh(D)-red cell membrane interactions. It is noteworthy that such a nonspecific mechanism has been proposed to contribute to the pathological deposit of cationic anti-DNA on the negatively charged membranes of kidney glomeruli during active SLE.8
The previous observation that most anti-Rh(D) IgGs are cationic5 and the present finding that the responsible a.a. residues are not clustered in CDRs raise the intriguing possibility that less cationic anti-Rh(D) may be produced in humans naturally or during Rh(D) immune responses. In the model described above, less cationic anti-Rh(D) would be difficult to detect or have low clinical significance since they may not be able to reach and bind the Rh(D) antigen in its negatively charged membrane environment.7 The eventual preparation of immunologically active and membrane-free recombinant Rh(D) antigen would permit to detect their reactivity. This hypothesis is supported by the functional characteristics of the Rh.D1 auto-anti-Rh(D) MoAb, which was prepared by phage display technology using papaı̈n-treated red cells.59 The Rh.D1 V region is coded by the rather anionic DP 71 VH5 (pI = 8.65) and IGLV3S1 Vλ (pI = 4.96) segments and the resulting V region pI (7.35) is consequently much lower than the average pI of our anti-Rh(D) V regions (9.44) (Table 3). Interestingly additional serological characterization showed that the Rh.D1 antibody was of low affinity with papaı̈n-treated red cells and unable to bind the Rh(D) antigen present on native untreated red cells.65 The hypothesis is also in agreement with the serological characteristics of some human sera. Indeed, weakly reactive natural ‘cold’ anti-Rh(D) have been found in the serum of some Rh(D)− persons1,66 and natural auto-anti-Rh(D) are frequently responsible for autoimmune hemolytic anemias.1 Biochemical characterization (pI) of these anti-Rh(D) would permit to determine if the nonclinically relevant and weakly reactive anti-Rh(D) are less cationic than immune anti-Rh(D) as observed with the Rh.D1 auto-anti-Rh(D) and if there is a shift towards a more cationic charge during the development of AIHA as recently observed with pathologic anti-DNA during active SLE.8
ACKNOWLEDGEMENT
We thank Drs André Darveau and Renée Bazin for helpful suggestions, Dr Roger Perreault for inspiring discussions on anti-Rh(D) serology, Dr Louis Thibault for advice and critical reading of the manuscript, Dr Catherine Collins for statistical analysis, and Samantha Hayward and Nicole Laurin for excellent secretarial support.
Supported by a grant from the Canadian Red Cross R & D Program.
Address reprint requests to Réal Lemieux, PhD, Canadian Red Cross Society, Blood Services, 1800 Alta Vista Dr, Ottawa, Ontario KlG 4J5, Canada.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal