Abstract
Human T-cell leukemia virus type I (HTLV-I)-infected T cells expanded in vitro by single-cell cloning provide a unique system for investigating virus-cell interactions in nonimmortalized T cells. By analysis of clones generated randomly from the blood of virus carriers, we confirm that CD4 T cells are the major reservoir of HTLV-I in vivo and show that most infected cells contain a single integrated provirus. Contrary to the situation in HTLV-I immortalized cell lines, the HTLV-I provirus was found to be transcriptionally silent in a high proportion of randomly generated T-cell clones and could not be reactivated by mitogenic stimulation. The spontaneous proliferation previously documented in HTLV-I–infected T-cell clones was not observed in silently infected cells, and therefore correlates directly with the expression of tax and other viral genes. The only cytokine mRNA found to be significantly elevated in the virus-producing clones was interleukin-6; however, receptor-blocking experiments argue against a role for IL-6 in the virus-induced cell proliferation. We observed a striking variation in the ability of individual HTLV-I–producing clones to immortalize fresh peripheral blood lymphocytes. This ability did not correlate with the levels of viral mRNA expression, gag p24 production, spontaneous proliferation, or tax-transactivation, possibly suggesting a role for host cell factors as determinants of viral infectivity or immortalization. Studies to elucidate the basis of this phenotypic heterogeneity should enhance our understanding of viral spread and pathogenesis.
THE HUMAN T-CELL leukemia virus type I (HTLV-I) is a T-cell tropic retrovirus that is endemic in southern Japan, the Caribbean, parts of Africa, and much of South America. HTLV-I is the etiologic agent of adult T-cell leukemia/lymphoma (ATLL), a malignant expansion of HTLV-I – infected T lymphocytes that occurs in approximately 1% of virus carriers.1 HTLV-I is also associated with a chronic progressive myelopathy (tropical spastic paraparesis/HTLV-I associated myelopathy [TSP/HAM]), uveitis, arthropathy, and polymyositis.2-6 In vitro, HTLV-I–infected T cells are characterized by increased expression of activation markers, spontaneous proliferation,7-9 and occasional immortalization.10-12 Freshly isolated peripheral blood T cells from HTLV-I–infected individuals exhibit an increased frequency of CD25 or HLA DR-expressing cells, suggesting that activation of T cells by HTLV-I does occur in vivo.8,13,14 The capacity of HTLV-I to activate and induce the proliferation of T cells in vivo is very likely central to its pathogenic potential. The increased replication rate of HTLV-I–infected cells may, for example, be a key factor predisposing these cells to secondary genetic alterations and eventual malignancy. The inappropriate activation and expansion of (autoreactive) T cells could also underlie the inflammatory disorders associated with HTLV-I infection.16
T-cell activation by HTLV-I has been attributed largely to the viral tax gene. The tax gene encodes a transcriptional transactivator, which is required for efficient use of the viral long terminal repeat (LTR) promoter,17 but which also activates the transcription of many cellular genes involved in cell growth and proliferation. Among others, these include early response genes (c-fos, junD, erg1, and erg2),18,19 cytokine growth factors (interleukin-2 [IL-2], IL-6, granulocyte-macrophage colony-stimulating factor [GM-CSF], tumor necrosis factor-α [TNF-α]) (reviewed in Buckle et al20 ), the α subunit of the IL-2 receptor (IL-2Rα),21,22 and the cell cycle regulator p21waf/cip1.23 The tax gene can act as an oncogene when expressed constitutively or at high levels and appears to be the only viral gene required for the immortalization of primary human T cells in vitro.24,25 Despite the extensive list of cellular genes known to be regulated by tax, the key molecular events underlying the induction of T-cell proliferation and in vitro immortalization by HTLV-I have yet to be determined. Autocrine IL-2 stimulation has been proposed as one mechanism of cell growth,21 but IL-2–independent proliferation has also been documented in both transformed and nontransformed T cells infected with HTLV-I.26-29
Much has been learned about interactions between HTLV-I and the host cell from the study of HTLV-I immortalized cell lines. These are easy to cultivate and produce large amounts of virus, but are not, however, representative of the infected cells found in vivo. By their nature, the immortalized lines have lost the normal control mechanisms governing cell growth and proliferation and are additionally characterized by frequent chromosomal rearrangements or mutations.23 30 HTLV-I–infected T cells expanded in vitro by single-cell cloning permit the analysis virus-cell interactions in cells that are closely related to peripheral blood T cells. In terms of their activation properties and growth, T-cell clones retain many of the characteristics of freshly isolated T cells and can be maintained in culture for extended periods by repeated mitogenic or antigenic stimulation. T-cell clones, therefore, represent a valuable system with which to investigate the molecular mechanism(s) underlying HTLV-I–induced T-cell proliferation.
We previously reported that 10% or more of T-cell clones generated randomly from the blood of subjects with HAM/TSP are infected with HTLV-I.26 31 The infected clones expressed elevated levels of IL-2Rα and were responsive to IL-2. Significantly, however, these clones were also able to proliferate in an IL-2–independent manner, a process termed “spontaneous clonal proliferation.”26 Here, a more detailed characterization of the HTLV-I–infected clones is presented. We examine the significance of viral gene expression, tax function and cytokine production in relation to the spontaneous proliferation of infected cells. We also demonstrate that not all virus-producing clones are able to immortalize fresh peripheral blood lymphocytes and begin to unravel factors contributing to proviral transmissibility and immortalizing potential.
MATERIALS AND METHODS
Generation and maintenance of T-cell clones.T-cell clones were generated as previously described32 by plating Ficoll-separated peripheral blood mononucear cells (PBMC) or cerebrospinal fluid cells (patient Ph only) at less than one cell per well of a 96-well V-bottom plate in the presence of 1 μg/mL phytohemagglutinin (PHA) and 105 feeder cells per well (irradiated allogeneic PBMC). Human IL-2 (Collaborative Biomedical Products, Becton Dickinson, Bedford, MA) was added 3 days later at 5% (vol/vol) (equivalent to 5 U/mL) and maintained continuously thereafter. Cells were grown in RPMI-1640 supplemented with 10% human AB serum, 5% IL-2, penicillin and streptomycin and maintained by restimulation with PHA and fresh feeder cells at 10- to 14-day intervals.32 To preserve the original growth characteristics of the cell, clones were maintained in this way for no more than 3 months, after which time, a fresh aliquot was thawed.
Identification of HTLV-I–infected clones.Clones were screened for HTLV-I proviral DNA by polymerase chain reaction (PCR) amplification with pol or X region primers as previously described.26,33 For detection of HTLV-II sequences, we used the taxI/II primers and conditions described by Lee et al.34
Phenotype determination.Cells were analyzed by flow cytometry after indirect staining with the following monoclonal antibodies (MoAbs): anti-Tac for CD25, Leu-3a for CD4, Leu-2a for CD8, Leu-4 for CD3, leu-M3 for CD14 (Becton Dickinson) as previously described.26
Spontaneous proliferation assay.Seven to 10 days after their last stimulation, cells were washed three times with phosphate-buffered saline (PBS), suspended in growth medium without IL-2 and plated in triplicate in a round bottom 96-well plate (105 cells per well). Where relevant, wells were supplemented with 10 U/mL rIL-2 (Boehringer Mannheim, Indianapolis, IN) or with the anti-gp130 MoAbs GPX7 and GPZ35 (a gift from TOSOH Corporation, Ayase-shi, Japan) at 10 μg/mL. After 72 hours, wells were pulsed with 1 μCi 3H-thymidine (DuPont NEN, Boston, MA) for 18 hours. Cells were then harvested and 3H-thymidine incorporation measured by liquid scintillation counting.
p24 antigen assay.Cell-free supernatants harvested from T-cell clones were stored at −20°C and gag p24 levels later measured using an enzyme immunoassay (EIA) (Coulter, Hialeah, FL). Assays were performed according to the manufacturer's instructions. The cut off for detection in this assay was 20 pg/mL.
Northern blot analysis.Total RNA was prepared from cells 6 days after PHA stimulation by single step acid guanidinium thiocyanate-phenol–chloroform extraction.35 For Northern blot analysis, 10 μg RNA was run on a 1% formaldehyde agarose gel, transferred to Zetaprobe membrane (BioRad, Hercules, CA), baked, and hybridized as described elsewhere.36 HTLV-I and glyceraldehyde-3–phosphate dehydrogenase (GAPDH) probes were prepared using a 9-kb Sac I fragment from pMT237 and a 1.27-kb rat GAPDH cDNA,38 respectively.
Southern blot analysis.Five-microgram samples of high-molecular-weight DNA were digested, electrophoresed in 0.7% agarose gels, and transferred to Zetaprobe membrane (BioRad) by capillary blotting in 0.4 mol/L NaOH. High-specific activity probes were prepared by the random priming method (Pharmacia oligo-labeling kit) and overnight hybridization was performed at 65°C in 0.25 mol/L sodium phosphate pH 7.4, 7% sodium lauryl sulfate containing 100 μg/mL sheared salmon sperm DNA. Membranes were washed in 3 × SSC, 0.1% sodium dodecyl sulfate (SDS) then 0.1 × SSC, 0.1% SDS at 65°C before autoradiography.
Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis of cytokine mRNAs.cDNA was prepared from total cellular RNA and analyzed by PCR as described.39 Primer sequences are shown in Table 1 and were designed to span several introns. Thermal cycling was done at 94°C for 1 minute, 55° or 60°C for 1 minute, and 72°C for 1.5 minutes for 28 to 35 cycles. Samples were normalized for their cDNA concentration by comparison of β-actin mRNA levels. In the cases of IL-2, IL-4, IL-6, and TNF-α, a semiquantitative technique was used whereby the test sample was spiked with known molar quantities of a competitor DNA fragment that is amplified with the same primers. PCR products were run on 2% agarose gels and the molar concentration of cDNA target sequences was deduced from the amount of competitor required to yield the same molar quantity of PCR product as the target cDNA.
PCR Primers
| . | . | . | 5′→3′ . |
|---|---|---|---|
| IL-1β | 358 bp | Forward | AGC ACT GAA AGC ATG ATC CGG GAC GTG GAG |
| Reverse | CAG ATA GAT GGG CTC ATA CCA GGG CTT GGC | ||
| IL-2 | 395 bp | Forward | TAC AGG ATG CAA CTC CTG TCT TGC ATT GCA |
| Reverse | GTT GCT GTC TCA TCA GCA TAT TCA CAC ATG | ||
| IL-4 | 365 bp | Forward | CTG CTA GCA TGT GCC GGC AAC TTT GTC CAC |
| Reverse | GAA GTT TTC CAA CGT ACT CTG GTT GGC TTC | ||
| IL-6 | 412 bp | Forward | GAC AAA CAA ATT CGG TAC ATC CTC GAC GGC |
| Reverse | AGT TGT CAT GTC CTG CAG CCA CTG GTT CTG | ||
| IL-10 | 352 bp | Forward | ATG CCC CAA GCT GAG AAC CAA GAC CCA |
| Reverse | TCT CAA GGG GCT GGG TCA GCT ATC CCA | ||
| IFN-γ | 355 bp | Forward | AGT TAT ATC TTG GCT TTT CA |
| Reverse | ACC GAA TAA TTA GTC AGC TT | ||
| TGF-β1 | 336 bp | Forward | GCC CTG GAC ACC AAC TAT TGC |
| Reverse | GCT GCA CTT GCA GGA GCG CAC | ||
| TGF-β2 | 417 bp | Forward | CCA GAA GAC TAT CCT GAG CCC GAG GAA GTC |
| Reverse | ATG TAG CGC TGG GTT GGA GAT GTT AAA TCT | ||
| TNF-α | 123 bp | Forward | TCT CGA ACC CCG AGT GAC AA |
| Reverse | TAT CTC TCA GCT CCA CGC CA | ||
| β-Actin | 406 bp | Forward | AAC CCC AAG GCC AAC CGC GAG AAG ATG ACC |
| Reverse | GGT GAT GAC CTG GCC GTC AGG CAG CTC GTA |
| . | . | . | 5′→3′ . |
|---|---|---|---|
| IL-1β | 358 bp | Forward | AGC ACT GAA AGC ATG ATC CGG GAC GTG GAG |
| Reverse | CAG ATA GAT GGG CTC ATA CCA GGG CTT GGC | ||
| IL-2 | 395 bp | Forward | TAC AGG ATG CAA CTC CTG TCT TGC ATT GCA |
| Reverse | GTT GCT GTC TCA TCA GCA TAT TCA CAC ATG | ||
| IL-4 | 365 bp | Forward | CTG CTA GCA TGT GCC GGC AAC TTT GTC CAC |
| Reverse | GAA GTT TTC CAA CGT ACT CTG GTT GGC TTC | ||
| IL-6 | 412 bp | Forward | GAC AAA CAA ATT CGG TAC ATC CTC GAC GGC |
| Reverse | AGT TGT CAT GTC CTG CAG CCA CTG GTT CTG | ||
| IL-10 | 352 bp | Forward | ATG CCC CAA GCT GAG AAC CAA GAC CCA |
| Reverse | TCT CAA GGG GCT GGG TCA GCT ATC CCA | ||
| IFN-γ | 355 bp | Forward | AGT TAT ATC TTG GCT TTT CA |
| Reverse | ACC GAA TAA TTA GTC AGC TT | ||
| TGF-β1 | 336 bp | Forward | GCC CTG GAC ACC AAC TAT TGC |
| Reverse | GCT GCA CTT GCA GGA GCG CAC | ||
| TGF-β2 | 417 bp | Forward | CCA GAA GAC TAT CCT GAG CCC GAG GAA GTC |
| Reverse | ATG TAG CGC TGG GTT GGA GAT GTT AAA TCT | ||
| TNF-α | 123 bp | Forward | TCT CGA ACC CCG AGT GAC AA |
| Reverse | TAT CTC TCA GCT CCA CGC CA | ||
| β-Actin | 406 bp | Forward | AAC CCC AAG GCC AAC CGC GAG AAG ATG ACC |
| Reverse | GGT GAT GAC CTG GCC GTC AGG CAG CTC GTA |
Metabolic labeling of cells and immunoprecipitation.A total of 5 × 107 cells stimulated 6 days earlier were washed with PBS and incubated for 12 hours in 5 mL cysteine-free RPMI-1640 containing 10% fetal calf serum, 5% IL-2, antibiotics, and 100 μCi 35S-cysteine per mL (DuPont NEN). The labeled cells were washed with PBS and solubilized in 1 mL lysis buffer (20 mmol/L Tris.HCl pH 8.0, 120 mmol/L NaCl, 0.2 mmol/L PMSF, 0.2 mmol/L sodium fluoride, 0.2 mmol/L EGTA, 5 μg/mL aprotinin, 0.2% sodium deoxycholate, 0.5% NP40) for 20 minutes on ice. Lysates were clarified by centrifugation at 33,000 rpm for 1 hour at 4°C, then divided into two equal parts. Viral antigens were immunoprecipitated directly from one part by incubation with 5 μL TSP patient serum coupled to protein A-Sepharose CL-48 beads (Pharmacia, Piscataway, NJ). The remaining lysate was incubated with lentil-lectin Sepharose 4B beads as described40 to enrich for glycoproteins before immunoprecipitating HTLV-I proteins with the TSP patient serum. Samples were washed five times with lysis buffer and run on a 7% SDS-polyacrylamide gel electrophoresis (PAGE) gel, which was fixed and treated with Enhance (DuPont NEN) before autoradiography.
Syncytium formation assay.T-cell clones stimulated 7 days earlier were mixed with the indicator cell line C8166-4541 at a 5:2 ratio. A total of 200-μL volumes of the suspension containing 1 × 105 clone cells and 4 × 104 C8166-45 cells were plated in duplicate in flat bottom 96-well plates. Eighteen hours later, wells were examined microscopically for the presence of syncytia. After gentle mixing, 100 μL of each cell suspension was cytospun, fixed for 15 minutes in methanol, and stained with Giemsa.
PBMC immortalization.Ficoll-separated PBMC (2 × 106 cells) from an HTLV-I negative donor were combined with an HTLV-I–infected T-cell clone (1 × 106 cells, day 11) and stimulated with 1 μg/mL PHA. Five percent human IL-2 was added 3 days later and maintained continuously thereafter. For infection by the MT2 cell line,10 2 × 106 PBMC were stimulated with PHA for 72 hours and cocultivated with 1 × 106 lethally irradiated (10,000 rads) MT2 cells in the presence of 5% IL-2. Cultures were fed with IL-2 at 3- to 5-day intervals and monitored for viability and cell number over a 4-month period.
Transient transfection of clones and measurement of chloramphenicol acetyltransferase (CAT) activity.A total of 2 × 107 cells stimulated 4 to 5 days earlier were transfected with pU3RI-CAT17 using the diethyl aminoethyl (DEAE) dextran method. Briefly, cells were washed with serum-free medium and incubated for 30 minutes with 5 μg DNA and 12.5 μg/mL DEAE dextran (Pharmacia). The cells were then washed and resuspended in IL-2–containing medium. Three days later, cytoplasmic extracts were prepared and CAT activity was measured as the percentage acetylation of 14C chloramphenicol in a 1-hour assay using 100 μg protein.42
RESULTS
Frequency and phenotype of HTLV-I–infected T-cell clones.Due to the high frequency of HTLV-I–infected cells and the noncytopathic nature of the infection, T cells infected with HTLV-I can readily be isolated from many carriers by direct single-cell cloning of peripheral blood T cells. The T-cell clones were obtained by culturing Ficoll-separated PBMC at less than one cell per well in the presence of PHA, feeder cells, and interleukin-2. With the exception of patient Pr, lethally irradiated PBMC from an allogeneic, HTLV-I negative donor were used as feeder cells to exclude the possibility of clones becoming infected in vitro. More than 140 clones have so far been generated from five subjects with HTLV-I associated myelopathy or polymyositis and from one healthy carrier. Between 11% and 33% of clones from the symptomatic subjects were found to be infected with HTLV-I (Table 2). This figure is in close agreement with previous estimates of proviral load based on Southern blot analysis of DNA extracted from PBMC,33,43,44 and suggests, as previously observed,45 that clones generated in this manner constitute a representative cross-section of peripheral blood T cells. HTLV-I–infected clones were initially identified by PCR screening for the presence of proviral DNA. Positive results were confirmed in all cases by Southern blot analysis of the integrated provirus (Fig 1). The greatest number of infected clones (33%) was obtained with patient Pr, but as autologous PBMC were used as feeder cells, some of these clones may have become infected in vitro. A lower frequency of infected clones (3%) was found in the asymptomatic carrier, and none of the 162 clones generated from an HTLV-II carrier was positive for taxII sequences. As quantitative studies have shown the virus load in HTLV-II carriers to be highly variable but frequently less than one copy per 100 PBMC,46-48 it seems probable that the absence of HTLV-II infected clones was a reflection of low virus load in this individual.
Frequency and Phenotype of Infected Clones
| Patient . | Virus . | Diagnosis . | Infected Clones/Total Clones . | Phenotype . | ||
|---|---|---|---|---|---|---|
| . | . | . | . | CD4 . | CD8 . | ND . |
| Pr | HTLV-I | TSP | 8/24 (33%)* | — | — | 8 |
| Du | HTLV-I | TSP | 7/40 (17%) | 5 | 1 | 1 |
| Mu | HTLV-I | TSP | 4/20 (20%) | 4 | 0 | — |
| Ph | HTLV-I | NSP | 1/9 (11%) | 0 | 1 | — |
| CP | HTLV-I | polymyositis | 3/20 (15%) | 3 | 0 | — |
| RM | HTLV-I | ASX | 1/30 (3%) | — | — | 1 |
| RB | HTLV-II | ASX† | 0/162 (<1%) | — | — | — |
| Patient . | Virus . | Diagnosis . | Infected Clones/Total Clones . | Phenotype . | ||
|---|---|---|---|---|---|---|
| . | . | . | . | CD4 . | CD8 . | ND . |
| Pr | HTLV-I | TSP | 8/24 (33%)* | — | — | 8 |
| Du | HTLV-I | TSP | 7/40 (17%) | 5 | 1 | 1 |
| Mu | HTLV-I | TSP | 4/20 (20%) | 4 | 0 | — |
| Ph | HTLV-I | NSP | 1/9 (11%) | 0 | 1 | — |
| CP | HTLV-I | polymyositis | 3/20 (15%) | 3 | 0 | — |
| RM | HTLV-I | ASX | 1/30 (3%) | — | — | 1 |
| RB | HTLV-II | ASX† | 0/162 (<1%) | — | — | — |
Abbreviations: ND, not determined; NSP, nonspastic paraparesis; ASX, asymptomatic carrier.
Autologous feeder cells were used.
Patient hospitalized with hepatitis.
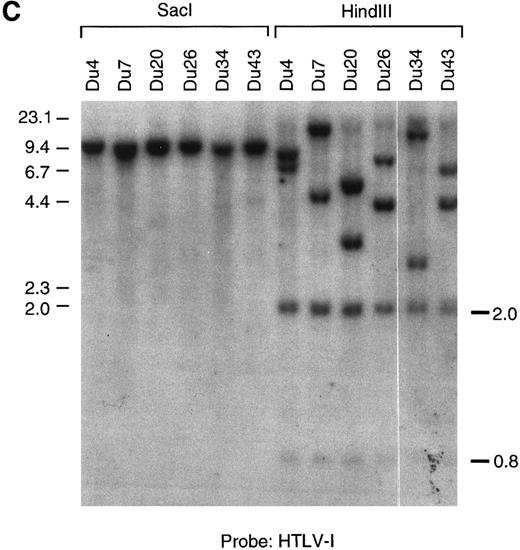
(A through C) Southern blot analysis of the HTLV-I provirus and T-cell receptor gene rearrangement in human T-cell clones. Genomic DNA digested with EcoRI (A, B) Sac I, or HindIII (C) was hybridized with the probe indicated. All clones except Du35 were infected with HTLV-I. (D) Restriction map of the HTLV-I genome. Numbers indicate the predicted length in kb of restriction fragments.
(A through C) Southern blot analysis of the HTLV-I provirus and T-cell receptor gene rearrangement in human T-cell clones. Genomic DNA digested with EcoRI (A, B) Sac I, or HindIII (C) was hybridized with the probe indicated. All clones except Du35 were infected with HTLV-I. (D) Restriction map of the HTLV-I genome. Numbers indicate the predicted length in kb of restriction fragments.
Of 11 HTLV-I positive clones analyzed, nine were CD4+, CD8− and two were CD8+, CD4−. The infected clones were morphologically similar to uninfected clones and did not have the multilobed nuclei typical of leukemic cells in ATL. The HTLV-I–infected clones were not immortal and required stimulation with PHA, feeder cells, and IL-2 at approximately 14-day intervals for continued growth. These results provide independent confirmation of the very high frequency of HTLV-I–infected cells in symptomatic carriers and also support our previous finding that the CD4 T lymphocyte subset forms the main reservoir of HTLV-I in peripheral blood.33
Infected T cells in blood are polyclonal and contain a single provirus.Genetic analysis has shown that fresh leukemic cells from ATL patients generally contain a single provirus, which may be incomplete,49,50 whereas HTLV-I–transformed cell lines cultured in vitro often contain multiple integrated proviruses51 (and references therein). T-cell cloning technology provides an opportunity to examine provirus copy number, integrity, and expression at the single-cell level in primary, nonimmortalized T cells. Southern blot analysis using an HTLV-I probe showed that most infected clones contain one integrated provirus. This was shown by the presence of a single hybridizing fragment when DNA was digested with EcoRI (which does not cut within the provirus) and also by the presence of two chromosomal junction fragments using HindIII (Fig 1). Two notable exceptions to this rule are the clones Ph1C and Mu16. Ph1C is a CD8 clone containing three discrete proviruses.52 Mu16 DNA gave a smear on EcoRI digestion. The EcoRI smear in Mu16 was not due to partial digestion or sample degradation as reprobing of the filter with a T-cell receptor gene probe gave two discrete bands (Fig 1B). These data are most consistent with the presence of multiple HTLV-I integration sites in the Mu16 clone, a hypothesis confirmed by genetic analysis of single-cell subclones (unpublished data, May 1992). Analysis of T-cell receptor gene rearrangement showed that all clones, including Ph1C and Mu16, were of single cell origin. No two clones so far tested from a single individual contained the same provirus integration site or T-cell receptor gene rearrangement, supporting previous reports that infected cells in the peripheral blood are largely polyclonal in origin (Fig 1B).44,53,54 This observation is easily reconciled with a report documenting the existence of clonally expanded virus-containing cells in the blood of carriers, as each expanded clone is thought to represent no more than 1% of the total T-cell population.15
Properties of HTLV-I–Infected Clones
| Clone . | Phenotype . | Spontaneous Proliferation (cpm) . | p24 (ng/mL) . | Syncytium Formation . | PBL Immortalization . | LTR-CAT Activity3-150 . |
|---|---|---|---|---|---|---|
| Du4 | CD4 | 8,172 | 8 | +++ | + | 10.3 |
| Du7 | CD8 | 15,525 | >28 | ND | ND | ND |
| Du26 | CD4 | 4,963 | 7 | − | − | 9.5 |
| Mu16 | CD4 | 6,912 | 10 | + | +++ | 13.3 |
| Du20 | CD4 | 218 | <0.02 | − | − | 0.65 |
| Du34 | CD4 | 182 | <0.02 | − | − | 0.74 |
| Du43 | CD4 | 148 | <0.02 | − | − | 0.51 |
| Mu40 | CD4 | 416 | <0.02 | − | ND | 0.36 |
| Ph1C | CD8 | 41 | <0.02 | − | ND | 0.45 |
| RM10 | ND | 180 | <0.02 | ND | ND | ND |
| CP9 | CD4 | 346 | <0.02 | ND | ND | ND |
| CP10 | CD4 | 198 | <0.02 | ND | ND | ND |
| Clone . | Phenotype . | Spontaneous Proliferation (cpm) . | p24 (ng/mL) . | Syncytium Formation . | PBL Immortalization . | LTR-CAT Activity3-150 . |
|---|---|---|---|---|---|---|
| Du4 | CD4 | 8,172 | 8 | +++ | + | 10.3 |
| Du7 | CD8 | 15,525 | >28 | ND | ND | ND |
| Du26 | CD4 | 4,963 | 7 | − | − | 9.5 |
| Mu16 | CD4 | 6,912 | 10 | + | +++ | 13.3 |
| Du20 | CD4 | 218 | <0.02 | − | − | 0.65 |
| Du34 | CD4 | 182 | <0.02 | − | − | 0.74 |
| Du43 | CD4 | 148 | <0.02 | − | − | 0.51 |
| Mu40 | CD4 | 416 | <0.02 | − | ND | 0.36 |
| Ph1C | CD8 | 41 | <0.02 | − | ND | 0.45 |
| RM10 | ND | 180 | <0.02 | ND | ND | ND |
| CP9 | CD4 | 346 | <0.02 | ND | ND | ND |
| CP10 | CD4 | 198 | <0.02 | ND | ND | ND |
Abbreviations: −, no syncytia; +, 10% to 30% of cells form syncytia; +++, >70% of cells form syncytia (on microscopic examination); ND, not done.
CAT activity expressed as percentage acetylation of 14C-chloramphenicol.
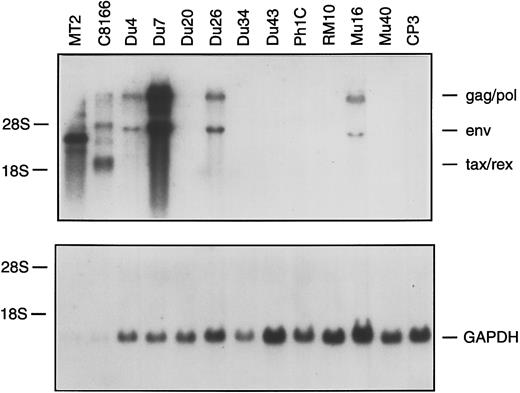
Northern blot analysis of total RNA from HTLV-I–infected clones. A total of 0.1 μg (MT2, C8166) or 10 μg (all other tracks) of total RNA was hybridized with an HTLV-I or GAPDH probe.
Northern blot analysis of total RNA from HTLV-I–infected clones. A total of 0.1 μg (MT2, C8166) or 10 μg (all other tracks) of total RNA was hybridized with an HTLV-I or GAPDH probe.
Silently infected clones occur at high frequency and do not proliferate spontaneously.We previously demonstrated that T-cell clones infected with HTLV-I show spontaneous proliferation, that is 3H-thymidine incorporation in the absence of IL-2.26,31 The spontaneous proliferation was not blocked by antibodies against the IL-2 receptor or by cyclosporin A, ruling out the possiblity of autocrine IL-2 stimulation.26 Of the 12 infected clones examined in this study, only four were found to proliferate spontaneously (Table 3). To examine the basis for this difference, we first compared HTLV-I gene expression in the 12 clones. Gag p24 was detectable at levels of 5 to >28 ng/mL in culture medium harvested from the four spontaneously proliferating clones, but not in the eight nonproliferating clones. This suggested a direct correlation between viral gene expression and spontaneous proliferation. Northern blotting of RNA from infected clones with an HTLV-I–specific probe showed the HTLV-I provirus to be transcriptionally silent in the p24-negative clones (Fig 2). Interestingly, two of the silently infected clones (Du20 and Du34) had been found to exhibit a low level of spontaneous proliferation when first isolated.31 We speculate that an initially low level of HTLV-I proviral expression in these clones has ceased completely in the course of in vitro culture.
The quantity of viral RNA in the productively infected clones was considerably lower than that found in two HTLV-I immortalized cell lines, C8166-45 and MT2 (Fig 2). By densitometry, the abundance of HTLV-I RNA in the Du4, Du26, and Mu16 cells was ≈100-fold lower than in the C8166-45 cell line (note that less RNA is loaded in the MT2 and C8166-45 tracks). The higher level of HTLV-I RNA present in the Du7 clone was still 5- to 10-fold lower than in C8166-45 cells. As a control, mRNA for glyceraldehyde-3-phosphate dehydrogenase was shown to be expressed at a similar level in all of the clones. In addition to establishing a strong correlation between HTLV-I expression and spontaneous proliferation, these findings illustrate a previously undescribed aspect of HTLV-I biology — the existence of a sizable fraction of circulating cells in which the provirus is transcriptionally inactive and could not be reactivated by mitogenic stimulation of the cell in vitro.
Cytokine mRNA expression by HTLV-I–infected and noninfected T-cell clones.Cytokines are known to play an important role in lymphocyte growth and survival and might be involved in the spontaneous proliferation of HTLV-I–producing clones. Previous studies have largely ruled out autocrine IL-2 stimulation as a mechanism,26 but the importance of other cytokines has yet to be determined. In this study, PCR was used to compare cytokine mRNA levels in a panel of productively infected, silently infected, and noninfected T-cell clones. The results are summarized in Table 4. As expected, tax mRNA was detected only in the four productively infected clones, whereas all clones were positive for β-actin mRNA. mRNAs for IL-1, IL-2, IL-4, IL-10, interferon-γ (IFN-γ), and TNF-α were detected in some or all of the clones, but the only cytokine mRNA showing a significant correlation with tax expression was IL-6. Normally produced by mononuclear phagocytes, fibroblasts, or endothelial cells, IL-6 secretion by HTLV-I–infected T cells has previously been documented55 and is attributed to tax-transactivation of the IL-6 promoter.56 57
RT-PCR Analysis of Cytokine mRNAs in T-Cell Clones
| Clone . | Productively Infected . | Silently Infected . | Noninfected . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Du4 . | Du7 . | Du26 . | Mu16 . | Mu16N4-150 . | Du34 . | Du43 . | Ph1C . | RM10 . | Du30 . | Du35 . | CP3 . |
| β-Actin | + | + | + | + | + | + | + | + | + | + | + | + |
| tax | + | + | + | + | ||||||||
| IL-1 | + | + | + | + | + | |||||||
| IL-24-151 | + | + | ++ | ++ | +++ | ++ | + | + | ++ | ++ | + | ++ |
| IL-44-151 | + | + | + | + | ++ | ++ | + | |||||
| IL-64-151 | ++ | + | + | + | ||||||||
| IL-10 | + | + | + | |||||||||
| IFNγ | + | + | + | + | + | + | + | + | + | + | + | + |
| TNF-α4-151 | + | + | + | + | ||||||||
| TGF-β1 | + | + | + | + | + | + | + | |||||
| Clone . | Productively Infected . | Silently Infected . | Noninfected . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Du4 . | Du7 . | Du26 . | Mu16 . | Mu16N4-150 . | Du34 . | Du43 . | Ph1C . | RM10 . | Du30 . | Du35 . | CP3 . |
| β-Actin | + | + | + | + | + | + | + | + | + | + | + | + |
| tax | + | + | + | + | ||||||||
| IL-1 | + | + | + | + | + | |||||||
| IL-24-151 | + | + | ++ | ++ | +++ | ++ | + | + | ++ | ++ | + | ++ |
| IL-44-151 | + | + | + | + | ++ | ++ | + | |||||
| IL-64-151 | ++ | + | + | + | ||||||||
| IL-10 | + | + | + | |||||||||
| IFNγ | + | + | + | + | + | + | + | + | + | + | + | + |
| TNF-α4-151 | + | + | + | + | ||||||||
| TGF-β1 | + | + | + | + | + | + | + | |||||
Subclone of Mu16.
Semiquantitative PCR. +, 0.001; ++, 0.01; +++, 0.1 attomoles of template cDNA in sample.
Receptor-blocking experiments were performed to investigate the possibility that the spontaneous proliferation of HTLV-I–producing clones is IL-6–mediated. Two MoAbs directed against the gp130 moiety of the IL-6 receptor and reported to neutralize the activity of IL-6,58 did not block the spontaneous proliferation (Table 5). Furthermore, neither recombinant IL-6 nor supernatant from HTLV-I–producing clones was able to induce the proliferation of noninfected T cells31 (and data not shown). Collectively, these results argue against a role for IL-6 in the virus-induced proliferation.
Effect of Anti-gp130 MoAbs on Spontaneous Proliferation of Du26 Cells
| IL-2 . | MoAb . | Proliferation (SEM) . |
|---|---|---|
| + | none | 63,728 (521) |
| − | none | 26,704 (981) |
| − | GPX7 | 26,678 (95) |
| − | GPZ35 | 26,304 (1,004) |
| IL-2 . | MoAb . | Proliferation (SEM) . |
|---|---|---|
| + | none | 63,728 (521) |
| − | none | 26,704 (981) |
| − | GPX7 | 26,678 (95) |
| − | GPZ35 | 26,304 (1,004) |
Spontaneous 3H-thymidine incorporation was measured after 48 hours incubation with the indicated MoAb.
Abbreviation: SEM, standard error of the mean.
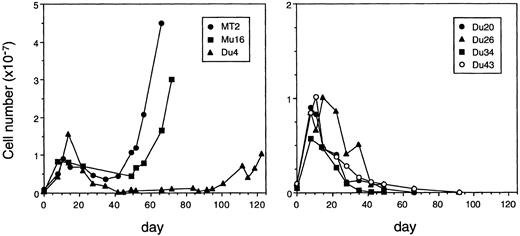
Variable immortalizing capacity of HTLV-I–infected clones.The dominant biological characteristic of HTLV-I in vitro is its capacity to immortalize primary lymphocytes. As none of the HTLV-I–producing clones described in this or previous reports are immortal, we used a cocultivation assay to determine whether the virus produced by these clones had the capacity to immortalize freshly isolated lymphocytes from a noninfected donor. PHA-stimulated PBMC were cocultured separately with six of the infected clones and also with lethally irradiated MT2 cells. The cultures were fed every 4 to 5 days with medium containing IL-2. As expected, cell numbers began to increase steadily in the MT2 cocultures after about 35 days. Of the three virus-producing clones examined, only Mu16 immortalized cells efficiently. The Du26 clone was not able to immortalize PBMC, and an immortal line emerged from the Du4 coculture at a late stage (Fig 3). The Du7 clone, which was harder to propagate, was not analyzed in this assay. Southern blot analysis of the T-cell receptor β gene rearrangement and proviral integration site in the Mu16- and Du4-immortalized lines confirmed these to be of PBMC and not clonal origin (data not shown). By flow cytometric analysis, the lines displayed the characteristic CD3+, CD4+, CD8−, CD25+ phenotype of HTLV-I–transformed cells (data not shown). Three nonproducer clones Du20, Du34, and Du43 all failed to give rise to immortalized cell lines (Fig 3).
Immortalizing capacity of HTLV-I–producing T-cell clones. Curves show the growth of PHA-stimulated PBMCs cocultivated with MT2 cells, with virus-producing T-cell clones (Mu16, Du4, Du26), or with silently infected clones (Du20, Du34, Du43).
Immortalizing capacity of HTLV-I–producing T-cell clones. Curves show the growth of PHA-stimulated PBMCs cocultivated with MT2 cells, with virus-producing T-cell clones (Mu16, Du4, Du26), or with silently infected clones (Du20, Du34, Du43).
Various viral and cellular parameters were examined in an attempt to explain the differing immortalizing capacity of the virus-producing clones. On repeated testing, the level of spontaneous proliferation in the Du26 clone was not significantly different from that of the Du4 or Mu16 clones, hence this property could not be correlated with the immortalizing potential of the clone. Viral mRNA and gag p24 levels were also comparable in the three clones (Fig 2 and Table 3). tax function was independently monitored in the infected clones by transiently transfecting cells with plasmid pU3RI-CAT, which contains a CAT reporter gene under the control of the HTLV-I LTR promoter. tax-dependent CAT activity could be demonstrated 72 hours later in the three virus-producing clones, but not in silently infected clones. There was no significant difference between the virus-producing clones with respect to the level of LTR transactivation (Table 3).
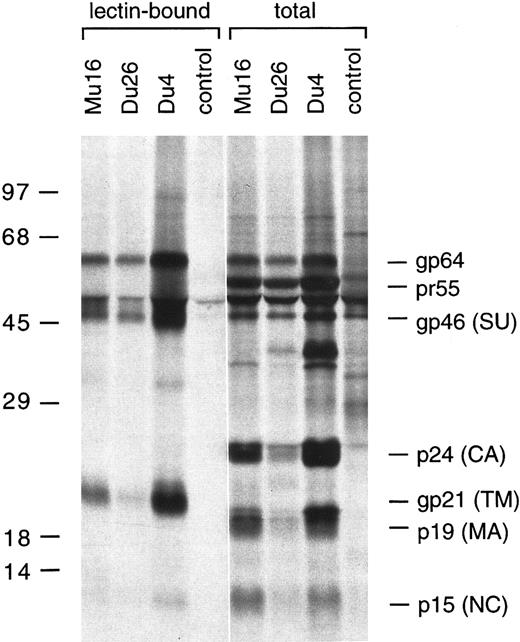
SDS-PAGE analysis of metabolically labeled HTLV-I proteins immunoprecipitated from the Du4, Du26, and Mu16 clones using a TSP patient serum.
SDS-PAGE analysis of metabolically labeled HTLV-I proteins immunoprecipitated from the Du4, Du26, and Mu16 clones using a TSP patient serum.
By immunoprecipitation and SDS-PAGE analysis, some differences in Gag and Env processing were observed, which may account for the variable immortalizing potential of the virus-producing clones. The envelope glycoproteins are synthesized as a 64-kD precursor, gp64, which is cleaved by a cellular protease to yield the mature surface glycoprotein, gp46 (SU), and a membrane-spanning subunit, gp21 (TM). gp64 was expressed in all three clones, but appeared to be cleaved very inefficiently in Du26 cells, as very little gp46 or gp21 was detected (Fig 4). gp64 cleavage was most efficient in the Du4 clone. As discussed below, inefficient processing of the Gag or Env precursors in Du26 cells may result in impaired virus infectivity, thereby explaining the lack of PBMC immortalization by this clone.
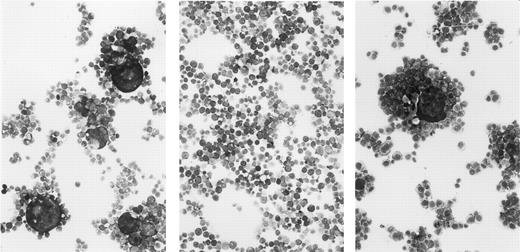
Lack of syncytium formation in a nonimmortalizing clone.A syncytium forming assay was used to examine whether a defect in envelope-mediated cell fusion might account for the failure of Du26 cells to immortalize PBMC. Cell fusion is observed when cells expressing HTLV-I envelope glycoproteins are mixed with cells bearing the virus receptor and results in the formation of multinucleated giant cells or syncytia.59 60 The molecular interactions between the envelope glycoprotein and its cellular receptor that lead to membrane fusion are believed to be closely related (or identical) to those involved in fusion of the viral and cellular membranes during virus entry. Hence, viruses that are unable to induce syncytium formation are generally also noninfectious.
The Du4, Du26, and Mu16 clones were analyzed for their ability to form syncytia when cocultured with C8166-45 indicator cells. Among the productively infected clones, Du4 was highly fusogenic, Mu16 less so, and no syncytia were observed with the Du26 clone (Fig 5). As expected, none of the silently infected clones formed syncytia. The syncytium-forming capacity of the three virus-producing clones correlated quite closely with the level of SU (gp46) expressed, being highest in the Du4 clone and absent in Du26. An essential role for gp46 in syncytium formation has been suggested by a report that env mutations which result in inefficient cleavage of the gp64 precursor also abolish syncytium formation.40 Host cell factors or env gene mutations that affect receptor binding may also contribute to the differing fusogenic capacity of the three clones. Whatever the basis for the fusion defect in Du26 cells, a low infectivity associated with this phenotype may account for the failure of this clone to immortalize PBMC.
Syncytium formation by HTLV-I–infected T-cell clones. The clones Du4 (left), Du26 (center), and Mu16 (right) were incubated overnight with C8166-45 indicator cells, cytospun, fixed, and stained with Giemsa.
Syncytium formation by HTLV-I–infected T-cell clones. The clones Du4 (left), Du26 (center), and Mu16 (right) were incubated overnight with C8166-45 indicator cells, cytospun, fixed, and stained with Giemsa.
DISCUSSION
Contribution of spontaneous proliferation to high virus loads.Previous studies have documented an extraordinarily high frequency of HTLV-I–infected cells in the PBMC of patients with HAM/TSP (≈10%) and a more variable, but occasionally high frequency (range, <0.1% to 6%) in asymptomatic HTLV-I carriers.33,43,44,61,62 HTLV-I is also characterized by an unusual degree of genetic stability, both within an individual carrier and when isolates from different geographical locations are compared.63,64 The high level of sequence conservation suggests that the high virus loads observed in vivo are achieved through the clonal expansion of infected cells rather than by multiple rounds of virus replication involving the inherently error-prone step of reverse transcription. The cell expansion hypothesis has recently received direct confirmation from a study in which PCR analysis of proviral integration sites showed the presence of clonally expanded T cells in the blood of healthy HTLV-I carriers.15 T-cell activation by the virus is likely to play a critical part in the in vivo proliferation of HTLV-I–infected cells and could occur by multiple mechanisms. Induction of the IL-2 receptor α chain mRNA expression (and therefore the high affinity IL-2 receptor) by tax, coupled with tax-mediated transactivation of the IL-2 promoter, could contribute to the expansion of infected cells via an autocrine IL-2 mechanism.21 The spontaneous proliferation shown by HTLV-I–infected T cells is likely to be an equally important mechanism that allows further expansion of the cells in the absence of IL-2. Two factors that will limit the extent of virus-driven cell proliferation in vivo are (1) a requirement for prior activation of the cell (to induce viral mRNA and protein synthesis), and (2) the elimination of viral antigen-positive cells by HTLV-I–specific cytotoxic T lymphocytes (CTLs), which are known to occur at high frequency in HAM/TSP patients.65,66 As a vast majority of the infected cells in peripheral blood do not express HTLVI,67 but can be induced to do so on mitogenic stimulation,68-72 we speculate that HTLV-I–induced cell proliferation most likely operates to enhance the vigor and duration of response in situations where an infected cell had been activated by some other mechanism.
As cytokines are known to play an important role in lymphocyte growth and might be involved in the virus-induced proliferation, we compared cytokine production by infected and noninfected T-cell clones. Interestingly, we found little evidence for overproduction of IL-1, IL-2, IL-4, and IFN-γ, which are reported to be tax-induced in certain cell types or in reporter gene assays.73-75 Our inability to confirm these findings in naturally infected clones suggests that these factors are not responsible for the virus-induced proliferation. Moreover, these data suggest that tax-mediated transactivation of these promoters may be cell-type dependent, require overproduction of the tax protein or alternatively require a combination of signals. In established T-cell lines, for example, tax synergizes with phorbol esters and mitogens in the induction of IL-2 and IFN-γ mRNA synthesis, but has little effect by itself on the expression of these mRNAs.21,22 73 The only factor showing clear cut differential expression in the presence of tax was IL-6. Despite its specific induction in virus-producing cells, receptor-blocking and other studies argue against a role for IL-6 in the spontaneous proliferation. Further studies are needed to elucidate the precise mechanism of virus-driven T-cell expansion, which can lead over time to malignancy or to the vast accumulation of infected cells observed in HAM/TSP patients.
Silent infection: The rule, not the exception.The infected clones described here were identified by screening randomly generated T-cell clones for the presence of proviral DNA, as opposed to screening for viral antigen production or for altered growth kinetics. An interesting finding to emerge from this approach was the high proportion of clones containing a transcriptionally inactive provirus. Transcription of the viral genome is known to be severely restricted in vivo,67 but can be induced in lymphocytes from ATL patients, HAM/TSP patients, and healthy HTLV-I carriers by mitogenic stimulation of the cells in vitro.68-72 By analysis at the single-cell level, we show here that mitogenic stimulation is not sufficient to reactivate provirus expression in a significant proportion of HTLV-I–infected T cells. These cells may contain a defective viral genome or alternatively be transcriptionally silenced by mechanisms that are not relieved by mitogenic stimulation. One such mechanism may be DNA methylation. Methylation of CpG dinucleotides has been shown to inhibit the transcription of many viral and cellular genes76 and has previously been demonstrated or suggested as a mechanism of HTLV-I latency.77-79 Another mechanism of transcriptional suppression involving the binding of cyclic AMP response element-binding protein (CREB) family members to a specific sequence within the R region of the HTLV-I LTR has been described and may be operative in the silently infected clones.80
HTLV-I–infected clones are not immortal.A notable feature of the HTLV-I–producing clones is that they are not immortalized. When compared with noninfected or nonproducer clones, the spontaneously proliferating cells have an extended but finite life span26 and die within 3 to 4 weeks in the absence of mitogenic stimulation. Constitutive HTLV-I expression may be required, but is clearly not sufficient for T-cell immortalization and secondary events, which result either in higher level tax expression or in the activation of cellular oncogenes, appear to be necessary for T-cell immortalization. We demonstrate, nevertheless, that virus produced by these T-cell clones is capable of immortalizing fresh PBMC in vitro. In light of the proven potential for T-cell transformation in these patients, the low incidence of ATL development in HAM/TSP patients, as well as other HTLV-I carriers, underscores the likely role of the immune system in restricting the transforming activity of HTLV-I in vivo. Interestingly, our in vitro studies show that the efficiency of PBMC immortalization by individual virus-producing clones was highly variable. Immortalizing capability did not correlate closely with viral mRNA or gag p24 levels, tax-transactivation, or spontaneous proliferation. Indeed, the Mu16 clone was almost as effective at immortalizing PBMC as the MT2 cell line, which produces 50 to 100 times more virus. The differences in immortalizing potential may be due to genetic variation within the HTLV-I genome or, alternatively, may reflect the influence of host cell factors on virus assembly and transmission. In either case, the existence of clonal, renewable populations with distinct phenotypes should allow the identification of viral or cellular factors, which influence this important phenotype.
CONCLUSION
The single-cell cloning system used in this study allows in vitro expansion and detailed molecular characterization of randomly selected T cells. This approach has allowed the detection of phenotypes not usually seen when HTLV-I–infected cells are selected on the basis of continuous growth, and we have uncovered evidence for frequent latency and occasional multiplicity of the integrated provirus. We demonstrate surprising heterogeneity with respect to virus fusogenicity and immortalizing capability, which are likely to have a direct bearing on pathogenic potential of the virus in vivo. The clonal nature of the system provides an opportunity to correlate directly such properties as latency, fusogenicity, and infectivity with sequence analysis of the proviral genome. Detailed investigation of virus-cell interactions in this system should provide a clearer understanding of the mechanisms governing virus-driven cell proliferation and may suggest strategies to limit the spread of the virus in vivo.
ACKNOWLEDGMENT
We thank Prof T. Kishimoto and TOSOH Corp, Japan, for the anti-gp130 antibodies and Dr J. Tooze for performing flow cytometric analyses.
Supported by the Sykes Trust, Harrogate, UK. P.H. was supported by the National Multiple Sclerosis Society (RG-2631-A-1), New York, NY.
Address reprint requests to J.H. Richardson, PhD, Division of Human Retrovirology, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal