Abstract
The Wiskott-Aldrich syndrome (WAS) is an X-linked recessive disorder described as a clinical triad of thrombocytopenia, eczema, and immunodeficiency. The gene responsible for WAS encodes a 502-amino acid proline-rich protein (WASp) that is likely to play a role in the cytoskeleton reorganization and/or in signal transduction of hematopoietic cells. However, the function and the regulation of the WAS gene (WASP) have not yet been clearly defined. We have studied WASP expression at the transcriptional level in freshly isolated mature peripheral blood cells and during hematopoietic development. For this purpose, we have isolated CD34+ hematopoietic precursor cells from cord blood. These cells were cultured in vitro with various growth factors to generate committed or mature cells belonging to different hematopoietic differentiation pathways, such as granulocytic (CD15+) cells, monocytic (CD14+) cells, dendritic (CD1a+) cells, erythroid lineage (glycophorin A+) cells, and megakaryocytic cells (CD41+). We have shown by reverse transcriptase polymerase chain reaction analysis that the WASP transcript is ubiquitously detectable throughout differentiation from early hematopoietic progenitors, including CD34+CD45RA− and CD34+CD45RA+ cells, to cells belonging to different hematopoietic lineages, including erythroid-committed and dendritic cells. In addition, Northern blot analysis showed that peripheral blood circulating lymphocytes (CD3+ and CD19+ cells) and monocytes express WASP mRNA. Several hematopoietic cell lines were tested and higher levels of expression were consistently detected in myelomonocytic cell types. By contrast, primary nonhematopoietic cells, including fibroblasts, endothelial cells, and keratinocytes, were consistently negative for WASP mRNA.
THE WISKOTT-ALDRICH syndrome gene (WASP) was recently isolated by positional cloning1 and was shown to be mutated in patients with Wiskott-Aldrich syndrome (WAS)1-4 or X-linked trombocytopenia.5-7 WAS is a severe, X-linked recessive disease characterized by thrombocytopenia, eczema, immune deficiency (reviewed in Remold-O'Donnell et al8 ), and increased incidence of malignancies.9 The immunologic abnormalities vary between patients9 and both signs of severe immunodeficiency10-13 as well as signs of hyperactivation of the immune system14-16 can be observed. Even after the identification of the gene responsible for WAS, the pathogenesis of this clinical syndrome remains unclear.
The WASP sequence encodes a proline-rich protein (WASp) whose function is still poorly defined. It has been shown recently that WASp may bind to Nck17 (an SH3/SH2 adaptor protein involved in signal transduction), to CDC42 Hs18-20 (a small GTPase involved in regulation of cytoskeleton formation), to the cytoplasmic tyrosine-kinase Fyn,21 and to the Src homology 3 domains of Btk, Itk, Tec, Grb2, and phospholipase C-γ,22 suggesting a role in lymphoid cell signaling. However, few data are yet available concerning the regulation of the WASP gene expression. In this study, we evaluated the expression of WASP at the transcriptional level during hematopoiesis. Analysis of X-chromosome inactivation in mature hematopoietic cells from WAS carriers has shown that mutations at the WASP locus cause a disadvantage in the survival and/or proliferation of those precursor cells that have the X chromosome with the mutant WASP allele as the active X. This selection process is already detectable in myeloid precursor cells23 and even in CD34+ hematopoietic progenitor cells.24 Based on these findings, it is possible that WASP is expressed throughout the hematopoietic development and functions in the survival/differentiation of all the hematopoietic cell lineages or, alternatively, that the gene defect is detrimental for the differentiation of hematopoietic precursors without being necessarily expressed in all hematopoietic cell lineages. After identification of WASP, it has been shown that the WASP transcript and WASp protein are in fact present in lymphoid1 and myelomonocytic cells.25 In this study, we present evidence that WASP expression is restricted to the hematopoietic system and show that WASP is ubiquitously expressed, at the transcriptional level, throughout hematopoietic differentiation from early progenitor cells to cells of different hematopoietic cell lineages at various stages of differentiation, including erythroid-committed and dendritic cells.
MATERIALS AND METHODS
Cell lines.The cell lines included human early erythroblastic cell lines (K562 and TF1), megakaryocytic cell lines (HEL, MO7E, and MV4), myelomonocytic cell lines (U937, THP1, JOSK1, and RCA 2), early myeloid progenitor cell lines (KG1 and KG1a), a T-cell line (MOLT4), a B-lymphoid cell line (CESS), and a myeloma cell line (IM-9). The following were used as nonhematopoietic cell lines: a fibroblast cell line (HFL), an epithelioid carcinoma (HELA), a melanoma cell line (FO-1), primary human umbilical vein endothelial cells (HUVEC; kindly provided by Dr B.Wolff, Sandoz, Vienna, Austria), and primary keratinocytes cultures (kindly provided by Dr G. Hinterhuber, Department of Dermatology, University of Vienna, Vienna, Austria). All cell lines were cultured in RPMI supplemented with 10% fetal calf serum at 37°C and 5% CO2.
Antibodies.The following murine monoclonal antibodies (MoAbs) were used in our study: CD34 (clone 8G12) and CD56 (clone Leu19) purchased from Becton Dickinson Immunocytometry Systems (Mountain View, CA); CD45RA (clone MEM 93) was kindly provided by V. Horejsi (Praha, Czech Republic); CD14 (clone MEM18) was from Ander Grub (Kaumberg, Austria); and CD15 (clone VIMD5), CD1a (clone VIT6b), CD41 (GPIIb/IIa; clone VIPL1), CD33 (clone 4D3), CD71 (clone VIP1), and glycophorin A (clone VIEG4) were produced in our laboratory.
Immunofluorescence staining procedure and cell sorting.Cells were incubated for 15 minutes at 0°C to 4°C with conjugated fluorescein isothiocyanate (FITC) or phycoerythrin (PE) MoAb; for biotin-conjugated primary antibodies, incubation with the second-step reagent streptavidin PE (Becton Dickinson) was applied. Subsequently, cells were washed and sorted by flow cytometry (FACS Vantage; Becton Dickinson).
Progenitors cell isolation.Cord blood samples were collected during normal full-term deliveries. Mononuclear cells were isolated using Ficoll/Hypaque (Pharmacia, Uppsala, Sweden) density gradient centrifugation. CD34+ cells were separated by magnetic cell sorting (CD34 isolation kit, MACS; Milteny Biotec, Bergish Gladbach, Germany) according to the manufacturer's instructions. The purity of the CD34+ population ranged from 87% to 98% (mean, 94%). The isolated CD34+ cells were stained with MoAb against CD34+ (PE) and sorted by flow cytometry (FACS Vantage; Becton Dickinson), or double surface stainings for CD34 (FITC) and CD45RA (PE) were performed and cells were sorted into CD34+CD45RA+ and CD34+CD45RA−.
Culture of CD34+ CB cells and cell sorting procedure.Isolated CD34+ cells were cultured in 24-well plates (2 to 5 × 104 cells in 1 mL/well) in RPMI supplemented with L-glutamine (2.5 mmol/L), penicillin (125 IE/mL), and pooled cord blood plasma (10%) at 37°C in an humidified atmosphere with 5% CO2. For growth induction, combinations of recombinant human cytokines were used. The basic cocktail contained recombinant human interleukin-3 (rhIL-3; 100 U/mL; Behring AG, Marburg, Germany), rhIL-6 (10 ng/mL; Sandoz, Basel, Switzerland), and recombinant human stem cell factor (rhSCF; 20 ng/mL; Amgen, Thousand Oaks, CA). Futher supplementation with recombinant human granulocyte colony-stimulating factor (rhG-CSF; 100 U/mL; Behring AG) and recombinant human granulocyte-macrophage colony-stimulating factor (rhGM-CSF; 100 ng/mL; Sandoz) was used to expand cells of the granulomonocytic lineage. The addition of thrombopoietin (kindly provided by Dr Dan Eaton, Genentech, Inc, South San Francisco, CA) at a concentration of 100 ng/mL to the basic cocktail resulted in the proliferation of megakaryocytic progeny. Erythroid-committed cells were obtained by culturing CD34+ cells with the basic cocktail additionally supplemented with 2 U/mL of recombinant human erythropoietin (rhEpo; Cilag, Schaffhause, Switzerland). Monocytes and dendritic cells were generated upon culture in the presence of GM-CSF, recombinant human tumor necrosis factor-α (rhTNFα; 50 U/mL; Bender, Vienna, Austria), and SCF. After 2 weeks in culture, standard staining and sorting procedures, as described above, were applied for the purification of selected populations: CD15+CD14−, CD41+CD33−, glycophorinA+CD71+, CD14+CD1a−, CD1a+CD14−.
Isolation of peripheral blood hematopoietic cells.Cell separations were performed as follows. Buffy coat from healthy volunteers was centrifuged on a Ficoll-Hypaque gradient (Pharmacia); the mononuclear cells were isolated from the interface. The monocyte population was obtained by plastic adherence. Subsequent incubation with anti-CD14–coated immunomagnetic beads (Dynal, Oslo, Norway) allowed a purification of greater than 96%. From the nonadherent fraction, T cells were retrived by incubation with anti-CD3 beads (Dynal); from the CD3− population, the CD19+ population (B lymphocytes) was obtained still with magnetic beads coated with CD19. The purity of the different populations was analyzed at the FACScan (Becton Dickinson) and was greater than 96%. The natural killer cells (NK) were sorted by flow cytometry using the surface marker CD56 in the population CD3−CD19−.
Northern blot analysis.RNA was isolated from cell lines and freshly isolated hematopoietic cells. Poly-A+ RNA (2 to 5 μg) was denatured in formaldehyde/formamide buffer and electrophoresed according to standard methods on a 2% agarose-formaldehyde gel. RNA was transfered on Duralon UV Membranes nylon filters (Stratagene, La Jolla, CA) and cross-linked by UV irradiation. Filters were prehybridized for 2 hours and hydridized for 20 hours at 42°C with probes labeled with 32P by random priming (GIBCO-BRL, Gaithersburg, MD). For WASP, a 750-bp cDNA probe was used from position 150 to 900 according to the sequence reported in Derry et al.1 For β actin, a 300-bp probe derived from polymerase chain reaction (PCR) amplification was used. After washing, blots were exposed to x-ray films at −70°C for 3 days.
Reverse transcriptase PCR (RT-PCR).Total cellular RNA was isolated from 5 × 104 sorted cells according to the RNeasy Total RNA kit (Qiagen, Hilden, Germany), and approximately 20 ng of total RNA was processed for reverse transcription. cDNA synthesis was performed with SuperScript II reverse transcriptase (GIBCO-BRL). One-half of the cDNA reaction was used as template for WASP amplification; one-tenth of the product was instead used for glyceraldehyde 3-phosphate dehydrogenase (G3PDH) amplification. The PCR reaction was performed in a total volume of 50 μL, 200 μmol/L of each dNTP, 25 pmol of each primer, and 0.5 U Taq DNA polymerase (Perkin Elmer Cetus, Norwalk, CT). DNA was denatured for 4 minutes at 94°C and amplified for 25 cycles (94°C for 20 seconds, 60°C for 30 seconds, 72°C for 60 seconds, extended for 5 minutes at 72°C, and stored at 4°C). The cDNA was amplified for only 20 cycles and the same PCR conditions for G3PDH. The primers used either for WASP or G3PDH amplification were designed with the computer program Oligo 4.0 (Primer Analysis Software, National Biosciences Inc, Plymouth, MN) using sequence data obtained from Genbank. For WASP RT-PCR amplification, the primers used were as follows: the upper primer at position 13, 5′ GAAGACAAGGGCAGAAAGCACC 3′; and the lower primer at position 899, 5′ GTTAGAGGTCTCGGCGTCGGT 3′. As an internal control for cDNA quality, we used at first β actin (data not shown) and subsequently G3DPH. For G3PDH PCR amplification, the primers used were as follows: the upper primer at position 869, 5′ TCAAAGGCATCCTGGGCTACA 3′; and the lower primer at position 1155, 5′ GAGGGGAGATTCAGTGTG GTG 3′. In all experiments, H2O was included as a negative control in the reverse transcriptase procedure and in the PCR amplification. The electrophoresed DNA was transfered to filter and hybridized as described above for Northern blot analysis with the respective probe. Blots were exposed to x-ray films for 3 to 10 hours.
RESULTS
WASP gene expression in hematopoietic and nonhematopoietic cell lines by Northern blot.In a first series of experiments, we studied WASP gene expression in a panel of hematopoietic and nonhematopoietic cell lines, and the results of a representative Northern blot are shown in Fig 1A. WASP transcript is present in the promyeloblastic leukemia KG1a, in the T-cell line MOLT4, in the B-lymphoblastoid cell line CESS, in the monocytic cell line U937, in the erythroblastic-megakaryoblastic leukemia cell line HEL92.1.7, in the erythroleukemia TF1 but not in a fibroblast cell line (HFL), in an epidermoid carcinoma cell line (A431), and in a primary culture of HUVEC. The WASP-indicated band has a size of approximately 2.0 kb. WASP transcript was also detected in other hematopoietic cell lines, including the megakaryoblastic leukemia MO7E and MOLM-1 cell lines, the myeloid leukemia KG1 and HL60 cell lines, and a myeloma cell line IM9 (data not shown). In contrast, all of the nonhematopoietic cell lines and primary cell cultures tested, including fibroblasts, endothelial cells, keratinocytes, or transformed cell lines (HELA [epitheloid carcinoma], FO-1 [melanoma cell line], and KATO [gastric carcinoma]), were negative for WASP mRNA (data not shown).
Northern blot analysis of poly-A+ RNA from various hematopoietic and nonhematopoietic cell lines and freshly isolated cells from buffy coat. (A) Five micrograms of poly-A+ RNA from the different cell lines (the B-lymphoblastoid cell line CESS, the erythroblastic-megakaryoblastic leukemia cell line HEL92.1.7, the promyeloblastic leukemia KG1a, the monocytic cell line U937, the erythroleukemia TF1, the T-cell line MOLT4, a fibroblast cell line [HFL], and the epidermoid carcinoma cell line [A431]) and from a primary culture of HUVEC was electrophoresed on a 2% agarose gel and subjected to blotting and hybridization as described in Materials and Methods. The hybridization with WASP probe (cDNA fragment from position 150 to 900) showed a major band of approximately 2.0 kb. Extended exposure did not show any WASP transcript in the endothelial cell line HUVEC, in the carcinoma A431, and in a fibroblast cell line (HFL). The blot was rehybridized with a β-actin probe for checking of uniformity loading. (B) Two micrograms of poly-A+ RNA from T cells (CD3+), B cells (CD19+), and monocytes (CD14+) was extracted and processed as described in (A).
Northern blot analysis of poly-A+ RNA from various hematopoietic and nonhematopoietic cell lines and freshly isolated cells from buffy coat. (A) Five micrograms of poly-A+ RNA from the different cell lines (the B-lymphoblastoid cell line CESS, the erythroblastic-megakaryoblastic leukemia cell line HEL92.1.7, the promyeloblastic leukemia KG1a, the monocytic cell line U937, the erythroleukemia TF1, the T-cell line MOLT4, a fibroblast cell line [HFL], and the epidermoid carcinoma cell line [A431]) and from a primary culture of HUVEC was electrophoresed on a 2% agarose gel and subjected to blotting and hybridization as described in Materials and Methods. The hybridization with WASP probe (cDNA fragment from position 150 to 900) showed a major band of approximately 2.0 kb. Extended exposure did not show any WASP transcript in the endothelial cell line HUVEC, in the carcinoma A431, and in a fibroblast cell line (HFL). The blot was rehybridized with a β-actin probe for checking of uniformity loading. (B) Two micrograms of poly-A+ RNA from T cells (CD3+), B cells (CD19+), and monocytes (CD14+) was extracted and processed as described in (A).
In conclusion, all of the hematopoietic cell lineages tested were positive in the Northern blot, but their levels of expression of WASP transcript were variable. In particular, lower levels of expression were detected in the erythroblastoid cell lines TF1 (Fig 1A) and K562 (data not shown), whereas higher levels of mRNA have been consistently observed in monocytic cell lines such as U937, JOSK1, and THP1. These data are in agreement with previous observations that the content of WASp may vary among different hematopoietic cell types.25
WASP gene expression in peripheral blood mature cells.To further characterize WASP gene expression, freshly isolated T cells (CD3+), B cells (CD19+), and monocytes (CD14+) obtained from the peripheral blood of healthy volunteers were purified by immunomagnetic separation techniques as described in Materials and Methods. As shown in Fig 1B, all of these cell populations express WASP mRNA. Interestingly, similar to what previously was observed in cell lines, monocytes show a higher level of WASP transcript as compared with T and B lymphocytes. Because the proportion of NK cells isolated was not sufficient for a Northern blot analysis, we used RT-PCR. Although our intent was primarily qualitative, we standardized the method to obtain a linear amplification. A low number of amplification cycles (25 for WASP RT-PCR) was used, followed by detection by Southern Blot. The data obtained after reverse transcriptase and PCR amplification of cDNA obtained from RNA of cell subsets used in the previous Northern blot are reported in Fig 2. In this experiment, we could show that the WASP is also expressed in NK cells (CD56+ cells; Fig 2, lane 4).
WASP expression by RT-PCR in T, B, and NK cells and in monocytes. Approximately 20 ng of the same RNA preparation (that was used in the Northern blot) was reverse transcribed with primers specific for WASP cDNA and PCR amplified (25 cycles). The PCR products were electrophoresed, blotted, and hybridized with the same WASP probe used in Northern blot analysis; the expected size of 900-bp is shown. For control of cDNA quality, PCR amplification with specific primers for the G3DPH sequence was performed (20 cycles) and shown by hybridization with a cDNA radioactive probe (a product of 300 bp). The lane with H2O represents the PCR-negative control.
WASP expression by RT-PCR in T, B, and NK cells and in monocytes. Approximately 20 ng of the same RNA preparation (that was used in the Northern blot) was reverse transcribed with primers specific for WASP cDNA and PCR amplified (25 cycles). The PCR products were electrophoresed, blotted, and hybridized with the same WASP probe used in Northern blot analysis; the expected size of 900-bp is shown. For control of cDNA quality, PCR amplification with specific primers for the G3DPH sequence was performed (20 cycles) and shown by hybridization with a cDNA radioactive probe (a product of 300 bp). The lane with H2O represents the PCR-negative control.
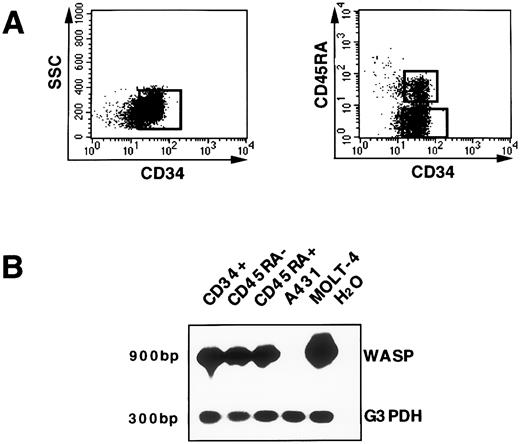
WASP gene expression in hematopoietic progenitors.CD34+ cells isolated from cord blood, as described in Materials and Methods, have been used as a model to investigate WASP gene expression at the transcriptional level in hematopoietic progenitor cells. Distinct developmental stages of human CD34+ progenitor cells can be identified by additional surface structures.26 Expression of the CD45RA antigen on CD34+ cells defines a population that is strongly enriched in committed granulomonocytic progenitors. By contrast, the CD34+CD45RA− subset defines early myeloid precursors and erythroid progenitors.27 28 Thus, we analyzed WASP mRNA in the total CD34+ population as well as in the CD34+CD45RA− and CD34+CD45RA+ subsets. FACS profiles indicating the strategy for sorting, after immunomagnetic enrichment, total CD34+ cells and CD34+CD45RA+, CD34+CD45RA− subsets are represented in Fig 3A. After cell sorting, only samples with a purity greater than 99% were further processed for RT-PCR analysis. WASP transcript was detected both in the bulk CD34+ population (lane 1) and in the CD34+CD45RA− (lane 2) and CD34+CD45RA+ (lane 3) subsets. Using the same number of cells (105) the T-cell line (MOLT-4) and the carcinoma cell line (A431) were run in parallel. No WASP transcript was shown in the A431 (lane 4); also, prolonged exposure of the autoradiograph did not show any positivity for WASP, similar to what was previously demonstrated by Northern blot analysis. Higher levels of WASP transcript were detected in the MOLT-4 cell line (lane 5) as compared with CD34+ cells. All of the cell lines that resulted to be negative for WASP mRNA in Northern blot were also tested by RT-PCR and no signal was identified (data not shown).
WASP gene expression in hematopoietic precursors. (A) The enriched CD34+ population isolated from cord blood was further purified by the cell sorting procedure as shown. In addition, CD34+ cells were stained with the CD45RA MoAb and the two subsets, CD34+CD45RA+ and CD34+CD45RA−, were selected, as represented. (B) RNA from CD34+, CD34+CD45RA+, and CD34+CD45RA− sorted cells; from the T-cell line MOLT-4 (used as a positive control for WASP amplification); and from the carcinoma cell line A431 (negative control for WASP expression) was reverse transcribed and subsequently amplified (25 cycles) either with primers specific for WASP cDNA that determine a PCR product of 900 bp or with primers specific for the G3PDH sequence (300 bp). The PCR products were shown by hybridization with the specific cDNA 32P-labeled probe. A WASP signal in A431 cell line was not detected even after extended autoradiographic exposure. The lane with H2O represents the PCR-negative control.
WASP gene expression in hematopoietic precursors. (A) The enriched CD34+ population isolated from cord blood was further purified by the cell sorting procedure as shown. In addition, CD34+ cells were stained with the CD45RA MoAb and the two subsets, CD34+CD45RA+ and CD34+CD45RA−, were selected, as represented. (B) RNA from CD34+, CD34+CD45RA+, and CD34+CD45RA− sorted cells; from the T-cell line MOLT-4 (used as a positive control for WASP amplification); and from the carcinoma cell line A431 (negative control for WASP expression) was reverse transcribed and subsequently amplified (25 cycles) either with primers specific for WASP cDNA that determine a PCR product of 900 bp or with primers specific for the G3PDH sequence (300 bp). The PCR products were shown by hybridization with the specific cDNA 32P-labeled probe. A WASP signal in A431 cell line was not detected even after extended autoradiographic exposure. The lane with H2O represents the PCR-negative control.
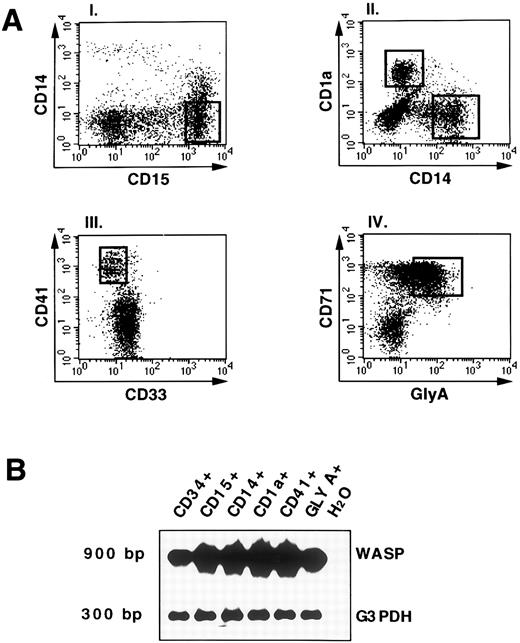
WASP gene expression in in vitro differentiated cells derived from CD34+ hematopoietic progenitors.To define the expression of WASP during hematopoiesis, freshly isolated CD34+ cord blood cells were cultured in the presence of different cytokine combinations as described in Materials and Methods. At 14 days of culture, cells were stained for the expression of lineage marker molecules. Figure 4A shows fluorescence stainings and sorting gates of cultured cells. The granulocytic population was defined as CD15+CD14− cells (Fig 4A, panel I). Cells of the monocytic and dendritic -c e l l l i n e a g e s w e r e i d e n t i f i e d a s C D 1 4 ;pl C D 1 a ;ms a n d -CD14−CD1a+, respectively (Fig 4A, panel II), as previously described.29 30 The marker molecule CD41 was used to define megakaryocytic progenies (Fig 4A, panel III); the erythroid progenitors were identified by coexpression of CD71 and glycophorin A (Fig 4A, panel IV). RT-PCR analysis of the different populations showed WASP expression in all of the cell lineages (Fig 4B) and, interestingly, even in committed erythroid cells.
WASP expression by RT-PCR analysis in differentiated and committed hematopoietic cell populations. (A) Representation of the staining and sorting gates of cultured cells. The culture of hematopoietic progenitors with various cytokines (as described in Materials and Methods) allowed the generation of different populations: the granulocytic sorted as CD15+CD14−; the monocytic and dendritic sorted, respectively, as CD14+CD1a− andCD1a+CD14−; the megakariocytes defined as CD41+CD33−; and the erythroid-committed, glycophorin A+CD71+ (GLY-A+). The open squares indicate the sorting regions. (B) RNA from FACS-sorted cell cultures (A) was processed as described and RT-PCR analysis was applied. PCR products for WASP (900 bp) or G3PDH (300 bp) were shown by hybridization as described above. As negative control, H2O was added instead of DNA in the PCR amplification.
WASP expression by RT-PCR analysis in differentiated and committed hematopoietic cell populations. (A) Representation of the staining and sorting gates of cultured cells. The culture of hematopoietic progenitors with various cytokines (as described in Materials and Methods) allowed the generation of different populations: the granulocytic sorted as CD15+CD14−; the monocytic and dendritic sorted, respectively, as CD14+CD1a− andCD1a+CD14−; the megakariocytes defined as CD41+CD33−; and the erythroid-committed, glycophorin A+CD71+ (GLY-A+). The open squares indicate the sorting regions. (B) RNA from FACS-sorted cell cultures (A) was processed as described and RT-PCR analysis was applied. PCR products for WASP (900 bp) or G3PDH (300 bp) were shown by hybridization as described above. As negative control, H2O was added instead of DNA in the PCR amplification.
DISCUSSION
Although the function of WASp is not yet fully defined, recent evidence suggests that it interacts with actin cytoskeleton organization18-20 and is also involved in signal transduction.17,21,22 So far, little is known about the expression and regulation of the WASP gene. The demonstration that carrier females of WAS exhibit a nonrandom pattern of X-chromosome inactivation in mature blood31,32 as well as in distinct hematopoietic precursors, including CD34+ cells,24 may either indicate that mutations at the WASP locus are detrimental at an early stage of differentiation or that selection against mutant cells continues throughout the differentiation process. To help solve this issue, we have studied WASP expression during hematopoietic differentiation. Stewart et al25 have previously reported that WASP mRNA is abundant in peripheral blood mononuclear cells and that different levels of expression are present in the hematopoietic cell lines MOLT-4, K562, and HEL92.1.7 and in one Epstein-Barr virus-transformed cell line; the development of MoAb against WASp allowed the authors to confirm these findings by analysis of WASp protein espression. We have extended the analysis at the transcriptional level by studing WASP expression during hematopoietic differentiation. We have included in our study primary and transformed hematopoietic as well as nonhematopoietic cell lines, peripheral blood subpopulations, cord blood-derived CD34+ hematopoietic progenitor cells, and committed hematopoietic cells obtained by culturing CD34+ cells in vitro with various growth factors. Our data show that the WASP transcript is ubiquitously present during hematopoietic cell differentiation. In particular, WASP transcript was consistently detected by RT-PCR in erythroid committed progenitors (glycophorin A+ CD71+) obtained by in vitro culture of purified cord blood CD34+ cells with appropriate cytokines. Analysis of X-chromosome inactivation in the erythroid cell lineage from carrier females of WAS performed by evaluating the expression of glucose-6-phosphate dehydrogenase isoenzymes in red blood cells had led to controversial results.33,34 Prchal et al34 had hypothesized that the gene defect in carriers of the Wiskott-Aldrich syndrome could be detrimental to committed hematopoietic precursors, including those belonging to the erythroid lineage. Our data clearly indicate that WASP is indeed expressed in erythroid-committed cells. On the other hand, no overt functional defects have been reported in red blood cells from WAS patients, suggesting that, in certain hematopoietic lineages, the defect may not be particularly detrimental and is possibly counteracted by other mechanisms.
We have also shown that high levels of WASP transcript are detectable in monocytes and dendritic cells. These data are in agreement with the previous demonstration by Stewart et al25 that the WASp protein is abundant in peripheral blood monocytes. Early studies by Blaese et al10 and Cooper et al12 suggested a defect in antigen processing. Whether this defect is due to impaired cytoskeleton reorganization and/or defective signaling and whether WASp protein is required for optimal interaction between antigen-presenting cells and T lymphocytes remain to be ascertained.
Finally, the ubiquitous expression of the WASP in hematopoietic cells, even at the progenitor cell level, and its complete absence in nonhematopoietic cells strengthens the hypothesis that this molecule might have an important functional role in hematopoietic differentiation. Moreover, the recent demonstration that a homologue of WASP is expressed in neuronal cells (N-WASP)35 indicates that WASp is a member of a novel class of cytoplasmic proteins that share a key role in cytoskeleton reorganization and signaling in distinct cell lineages.
ACKNOWLEDGMENT
The authors gratefully acknowledge Prof L.D. Notarangelo and Prof M. Eibl for critical reading of the manuscript and A. Renner for the FACS sorting operation.
Supported by Fonds zur Förderung der wissenschaftlichen Forschung in Österreich.
Address reprint requests to Ornella Parolini, PhD, Institute of Immunology-VIRCC at SFI, University of Vienna, Brunnerstrasse 59, A-1235 Vienna, Austria.

![Fig. 1. Northern blot analysis of poly-A+ RNA from various hematopoietic and nonhematopoietic cell lines and freshly isolated cells from buffy coat. (A) Five micrograms of poly-A+ RNA from the different cell lines (the B-lymphoblastoid cell line CESS, the erythroblastic-megakaryoblastic leukemia cell line HEL92.1.7, the promyeloblastic leukemia KG1a, the monocytic cell line U937, the erythroleukemia TF1, the T-cell line MOLT4, a fibroblast cell line [HFL], and the epidermoid carcinoma cell line [A431]) and from a primary culture of HUVEC was electrophoresed on a 2% agarose gel and subjected to blotting and hybridization as described in Materials and Methods. The hybridization with WASP probe (cDNA fragment from position 150 to 900) showed a major band of approximately 2.0 kb. Extended exposure did not show any WASP transcript in the endothelial cell line HUVEC, in the carcinoma A431, and in a fibroblast cell line (HFL). The blot was rehybridized with a β-actin probe for checking of uniformity loading. (B) Two micrograms of poly-A+ RNA from T cells (CD3+), B cells (CD19+), and monocytes (CD14+) was extracted and processed as described in (A).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/90/1/10.1182_blood.v90.1.70/5/m_bl_0023f1.jpeg?Expires=1769138569&Signature=qLRQIiXFfL72P~3ow~NYkzU~XBrfHemWoOAL~Mb8Ln26juanSVpnwO8YoxNMqzipdPu6fylCai~dlS6qhfOqLsYtsb75sM7DtfXYUj42u8jErQGcb7oQ56vtAMOgz8nnug-llWpfAKL~JW~pxZiz4EaOJB6zEpCbRuwDyyCA~W8wAGk50epymq9ZdAMxWkJuReORdqSCpUmnObEFxW7YfPTuCrP1w8fLAK0Y4hyTnl4dOIiYIytUc95IBjj9UA1vIozW3EtMBlasHDK2YLMDZ1ofOl3kEKmHJ7~ROlAtF4UUFjqrKzhXhBi8~MJJ5mQJOv5eNMiFn8-xT4dLjS-9Ew__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal